colistin resistant Klebsiella pneumoniae ST101 high-risk clone in Turkey
G ULS¸EN HAZIROLAN €
1pand ALPER KARAG OZ €
21Department of Medical Microbiology, Faculty of Medicine, Hacettepe University, 06100, Ankara, Turkey
2Department of Microbiology, Faculty of Molecular Biology and Genetics, Usak University, 64200, Usak, Turkey
Received: August 11, 2020 • Accepted: September 1, 2020 Published online: November 9, 2020
ABSTRACT
Carbapenemase-producing and colistin resistant Klebsiella pneumoniae has become a worldwide healthcare problem. This study describes molecular characterization of carbapenemase-producing and colistin resistant clinicalK. pneumoniaeisolates.
A total of 93 non-replicate carbapenem and colistin resistantK. pneumoniaewere recovered from clinical specimens in a university hospital during 2017–2019. Detection ofblaOXA-48,blaKPC,blaNDM-1, blaIMP,blaVIM-1and mcr-1,-2, -3,-4, -5,-6, -7,and-8genes was performed by PCR. The bacterial isolates were assigned to clonal lineages by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST).
All isolates harbored blaOXA-48 and only two isolates harbored blaOXA-48, and blaNDM-1 genes together. In colistin resistantK. pneumoniae,mcr-1was detected in two (2.1%) isolates. Ninety three isolates ofK. pneumoniaewere categorized into three clusters andfive pulsotypes. MLST revealed two different sequence types, ST101 (89/93) and ST147 (4/93).
In our study ST101 was found to be a significantly dominant clone carryingblaOXA-48and among our strains a low frequency ofmcr-1gene was determined. The emergence of colistin resistance was observed inK. pneumoniaeST101 isolates. ST101 may become a global threat in the dissemination of carbapenem and colistin resistance.
KEYWORDS
Klebsiella pneumoniae, carbapenemase, colistin resistance,mcr-1,blaOXA-48, PFGE, MLST, ST101, ST147
INTRODUCTION
Klebsiella pneumoniae is an opportunistic pathogen which can cause different types of healthcare-associated infections. Enhanced use of carbapenems in clinical practice, promoted emergence of carbapenem-resistantK. pneumoniae (CRKP) worldwide in recent years [1].
CRKP has mainly been link toK. pneumoniaecarbapenemase (KPC), oxacillinase-48 (OXA- 48), and metallo-b-lactamases (MBLs), such as NDM, IMP, and VIM type carbapenemases.
While these plasmid-encoded carbapenemases have been increasingly reported worldwide, their prevalence varies geographically [2]. The first identified OXA-48 producer was a K.
pneumoniaestrain isolated in Turkey in 2003 [3]. Since then, OXA-48 producers have been extensively reported from Turkey as a source of nosocomial outbreaks [4–8]. Worldwide distribution of OXA-48 now includes countries in Europe, in the southern and eastern part of the Mediterranean Sea, and Africa [3–8].
Treatment of infected patients with CRKP is always problematic due to their multidrug resistance phenotype, and several therapeutic options have been considered. Colistin is one of these therapeutic options. However, colistin resistance had been observed in CRKP isolates,
Acta Microbiologica et Immunologica Hungarica
67 (2020) 4, 216–221 DOI:
10.1556/030.2020.01275
© 2020 The Author(s)
RESEARCH ARTICLE
*Corresponding author.
Tel.:þ90 312 305 1560.
E-mail:drgulsencetin@yahoo.com
and rapid dissemination of colistin resistant isolates has been recently reported [9]. Colistin resistance mechanisms are presumed to be linked to chromosomal mutation untransferable via horizontal gene transfer [10]. Recently, several plasmid-mediated colistin resistance genes, named mcr, encoding pEtN transferases, have also been reported in K. pneumoniae[11].
Dissemination of CRKP is mainly caused by the spread of a few successful clones. Major representatives of these high-risk clonal lineages include sequence type (ST) 11, ST15, ST307, ST17, ST37, ST101, and ST147 strains [1].
ST258 strains are major players in the worldwide spread of KPC-type carbapenemases, and are responsible for 68% of the CRKP outbreaks [12]. ST101 strains harbor different clinically-relevant resistance determinants, such as carba- penemases of the KPC, OXA-48, VIM, and NDM types [1].
In this study, carbapenemase producing and colistin resistant clinical K.pneumoniae isolates were characterized to evaluate genetic differences and relationships, and prev- alence of carbapenem resistance determinants, as well as to determine plasmid-mediated colistin resistance mechanism.
MATERIAL AND METHODS
Bacterial isolates and susceptibility testing
We retrospectively analyzed ninety-three carbapenem and colistin resistant K. pneumoniae isolates consecutively iso- lated from patients who were hospitalized at the Hacettepe University Hospitals between October 2017 and December 2019. The Hacettepe University Hospitals are tertiary care hospitals of 1,040 beds that provides specialized attention to a population size of∼5.504 million habitants in the capital of Turkey. In the period between October 2017 and December 2019, altogether 8624 K. pneumoniae isolates were obtained from routine microbiological cultures of clinical samples. In total, 93 carbapenem and colistin resis- tant K. pneumoniae isolates were obtained. These isolates were detected from blood (n534), urine (n526), abscess (n513), tracheal aspirate (n511), peritonealfluid (n54), cerebrospinal fluid (n5 2), synovialfluid (n 5 1), pleural fluid (n 5 1), and pericardial fluid (n 5 1). Isolates were identified with conventional tests (Gram staining, catalase and oxidase tests), and matrix assisted lazer desorption ionization time of flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Germany). All isolates were identified with a score≥2.0; accurate identification to the species level by MALDI-TOF MS. Antibiotic susceptibility profiles of isolates were determined by BD PhoenixÔ automated sus- ceptibility testing system (Becton Dickinson and Company BD, USA). Isolates non-susceptible to at least one carbape- nem (ertapenem, meropenem, and imipenem) were also tested for carbapenem resistance by gradient test (bioMerieux, France) according to manufacturer’s instructions. CRKP was defined when the isolate was resistant to ertapenem, mer- openem or imipenem by gradient test. Colistin MICs were determined for 93 CRKP isolates using the SensititreÔplate
(Thermo Fisher, UK). Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoints [13].Escherichia coliATCC 25922 andE.
coliNCTC 13846 (mcr-1 positive) was used for quality con- trol. The isolates were stored in tryptic soy broth containing 10% (v/v) glycerol at808C until use.
Molecular analysis of carbapenem and colistin resistance
Genomic DNA was isolated using the QIAsymphony DSP Virus/Pathogen kit in the QIAsymphony system according to the manufacturer’s instructions (Qiagen, USA). OXA-48, KPC, NDM-1, IMP, and VIM-1 carbapenemases were identified by PCR amplification and sequencing as described previously [14]. The colistin resistant isolates were screened by simplex PCRs for the presence of mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, mcr-6, mcr-7, and mcr-8 genes [15–21]
(Supplementary Table S1).
PFGE
Pulsed-field gel electrophoresis (PFGE) was performed as per a method described in a previous study [22]. A thin slice of plug was digested overnight at 37 8C with 50 U of the XbaI restriction enzyme according to the manufacturer’s instructions. The restriction fragments were separated through PFGE in 1% agarose gel (Bio-Rad, USA) with 0.5x TBE buffer (45 mM Tris, 45 mM boric acid, and 1.0 mM EDTA [pH 8.0]) for 22 h at 200 V and 148C, with ramp times of 2 s-40 s using the CHEF Mapper apparatus (Bio- Rad, USA). The gels were stained in ethidium bromide (1 mg/mL), were viewed under an ultra-violet trans- illumination. Digital images were stored electronically.
PFGE banding patterns were analysed with the BioNumerics Software (Applied Maths, Belgium) using the dice coefficient and unweighted pair group method with arithmetic averages algorithm. PFGE patterns were compared and analysed ac- cording to the criteria mentioned by Tenover et al. [23].
MLST
Multilocus sequence typing (MLST) was performed on K.
pneumoniaeisolates according to the protocol described on theK. pneumoniaeMLST website (https://bigsdb.pasteur.fr/
klebsiella/klebsiella.html). Seven conserved housekeeping genes (gapA,infB,mdh,pgi,phoE,rpoB, andtonB) were used [24]. MLST results were typed according to the updated internationalK. pneumoniaeMLST database at the Pasteur Institute in Paris, France.
RESULTS
Bacterial isolates and susceptibility testing
In the period between October 2017 and December 2019, a total of 8,624 clinical isolates ofK. pneumoniaewere isolated from patients admitted to Hacettepe University Hospitals.
Among those, 2,259 isolates (26.2%) were non-susceptible to
at least one carbapenem by gradient test and were tested for colistin resistance. A total of 93 isolates (4.1%) out of this subset showed a colistin resistant phenotype by Sensititre with minimum inhibitory concentrations (MIC) that ranged between 4 and 128
m
g/m. Carbapenem and colistin MIC ranges, MIC50 and MIC90 profiles of the isolates are sum- marized inTable 1. Overall carbapenem and colistin resis- tance rates are shown in Supplementary Table S2.Molecular analysis of carbapenem and colistin resistance
The PCR results indicated thatblaOXA-48gene was detected in allK. pneumoniae isolates (100%), and 2.1% (n5 2) of the isolates co-produced blaOXA-48, and blaNDM-1. Other tested carbapenemase genes, such as blaKPC, blaIMP, and blaVIM-1could not detected in any of the isolates. Detection of mcr 1-8 genes using PCR technique revealed that two (2.1%) isolates were positive formcr-1. Nomcr-2, -3,4, -5, -6, -7,and -8were detected among all tested isolates.
PFGE
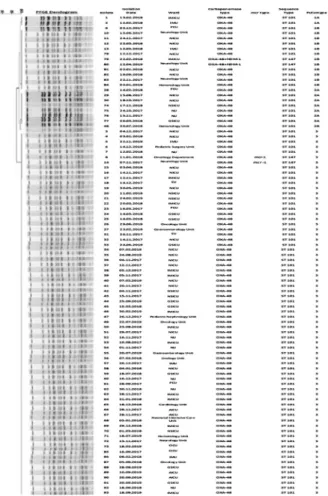
The characteristics of the molecular epidemiology of the 93 carbapenem and colistin resistantK. pneumoniaeisolates are displayed in Fig. 1. All the 93 carbapenem and colistin resistant K. pneumoniae isolates were grouped into three cluster andfive pulsotypes. Cluster three is the largest cluster that possesses 71 isolates. Fourteen isolates belonged to cluster one and eight isolates belong to cluster two. PFGE discriminatory power was of 96%, as calculated by Simp- son’s Index of Diversity [25].
MLST
Ninety-three carbapenem and colistin resistantK. pneumo- niaeisolates were analysed by MLST, and two ST types were detected. ST101(95.6%, 89/93) was the dominant ST type followed by ST147 (4.4%, 4/93). Among four isolates of
ST147, two isolates carried both blaOXA-48 and blaNDM-1 genes, and the other two ST147 clones coproduceblaOXA-48 andmcr-1genes (Fig. 1).
DISCUSSION
Multidrug resistant pathogens have become a global prob- lem recently [12]. K. pneumoniaeis an important nosoco- mial multidrug resistant pathogen that can cause high morbidity and mortality [26]. After widespread dissemina- tion of carbapenemase producing K. pneumoniae isolates;
colistin resistance has emerged in K. pneumoniae isolates and caused problems in treatment modalities [9, 26, 27].
CRKP isolates that produce carbapenemases such as the OXA-48, KPC, VIM-1, NDM-1, and IMP, have been re- ported worldwide [1, 3, 5–7, 9, 10, 26]. KPCs are the most clinically common enzymes, and have been detected in North America (especially the United States), South Amer- ica (Colombia, Argentina), Europe (Greece, Italy, Poland), Asia (China), and the Middle East (Israel) [28–30]. Turkey is a country with a specific epidemiology, where OXA-48 carbapenemase has been extensively identified. However, Table 1.MIC (mg/mL) profiles of carbapenem and colistin resistant
K. pneumoniaeisolates
2017 (n518) 2018 (n538) 2019 (n537) Colistin
MIC range 4–64 4–128 4–64
MIC50 4 4 4
MIC90 16 32 64
Meropenem
MIC range 16–128 16–128 16–256
MIC50 16 16 16
MIC90 32 64 128
Imipenem
MIC range 16–64 16–128 8–64
MIC50 16 16 8
MIC90 32 64 32
Ertapenem
MIC range 2–64 2–128 2–64
MIC50 2 16 16
MIC90 32 32 32
Fig. 1.Dendrogram based on pulsed-field gel electrophoresis pattern analysis (PFGE) of 93 colistin and carbapenem resistant
K. pneumoniaeisolates from different wardss and their ST determined via multilocus sequence typing (MLST)
first KPC-2-positiveK. pneumoniaehave been identified in Turkey, in 2014 [31]. Since then, sporadic KPC-producing K. pneumoniaewas reported [32, 33]. In the present study, we didn’t detectblaKPCgene among tested carbapenem and colistin resistant K. pneumoniaeisolates.
MBLs identified inK. pneumoniae mainly reported from Japan (IMP), Taiwan (IMP), Indian subcontinent (NDM), Balkan states (NDM), and Greece (VIM) [33]. Recentfindings suggest that the Balkan states and the Middle East may act as secondary reservoirs of NDM-1 producers. In the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) study, Turkey was classified as a stage 3 country on a scale of 1–5 (1: no reported case; 5: endemic situation) for the existence of NDM carbapenemases. Among CRKP, NDM-1 carbapenemases detected between 6.3% and 52.9% in our country [34, 35]. Moreover, the coproduction of both OXA-48 and NDM-1carbapenemases has been frequently reported. We detected two (2.1%) NDM-1-producing isolates, which have already been found to harbour OXA-48 carba- penemases. Interestingly, NDM-1 carbapenemase detected in a very low rate compared to our country results. Our hospital setting is not an endemic region for blaNDM-1 positive K. pneumoniae.The carbapenemase genesblaIMPandblaVIM were reported in a low but notable incidence in Turkey like other countries [27, 35–38]. In our study,blaIMPandblaVIM genes were not detected in any of the isolates.
OXA-48-producing K. pneumoniae was first reported from Turkey and it is endemic for our country [30, 32, 36, 37]. The emergence of the OXA-48 enzyme is mediated by the rapid spread of a broad host-range conjugative plasmid harboring the blaOXA-48 gene. Plasmid harboring blaOXA-48 with the Tn1999.2 transposon detected from a K. pneumoniae isolate in Turkey [39]. OXA-48-producing K. pneumoniaeis also endemic in certain North African and European countries (Morocco, Tunisia, Spain, Belgium) [40]. OXA-48-producing K. pneumoniae remain relatively uncommon in the United States and Canada [41]. As it was expected, we detected blaOXA-48 gene in all tested CRKP isolates. However, two of these isolates were positive for both blaNDM-1andblaOXA-48genes.
Multilocus sequence typing is an excellent method in evolutionary studies for exploring the common ancestral lineages of bacterial isolates [20]. Various ST types (ST11, ST14, ST101, ST147, and ST258) and resistance mechanisms can be related to carbapenem and colistin resistance in K.
pneumoniae[41]. A singleK. pneumoniaeclone ST258 was identified extensively worldwide, indicating that it may have contributed to the spread of the blaKPCgenes [26]. On the other hand, KPC-producingK. pneumoniaeremains rare for our country [31, 32]. Among all tested isolates, we didn’t detect KPC-producingK. pneumoniaeand also its emerging high-risk clone ST258. We found that the epidemicKlebsi- ella pneumonia isolate in our hospital was in ST101 type.
ST101 was previously accepted as a high-risk epidemic clone, and it was reported that the ST101 clone was asso- ciated with various b-lactamases, including NDM-1, OXA- 48, and CTX-M-15 [38]. Nevertheless, CRKP assigned to ST101 are carbapenem resistant frequently because of the
production OXA-48. David et al., analysed the genome se- quences ofK. pneumoniaestrains, isolated from patients in 244 hospitals in 32 countries during the European Survey of Carbapenemase-Producing Enterobacteriaceae. CRKP are concentrated in four clonal lineages, ST11, ST15, ST101, ST258/512, and authors identified OXA-48-producing ST101 clones in Romania, Spain, and Turkey [39]. Also, the emergence of colistin resistance has been observed in CRKP isolates, and colistin resistance was shown to be associated with the ST101 clone. Detection of carbapenem and colistin resistantK. pneumoniaeST101 was reported from Italy and Serbia [42, 43]. A large multicentre cohort study, describe the molecular characteristics of clinical colistin and carba- penem resistant K. pneumoniae isolates. Researchers observed a significant association between ST101, OXA-48, and colistin resistance [44]. Our study reports the clonal expansion of emerging ST101 clone associated with OXA-48 producing and colistin resistance in our hospital settings.
K. pneumoniaeST147 is an emerging high-risk clone that wasfirst identified in Greece and has been associated with VIM and KPC carbapenemases in that country [45]. This global ST has also been associated with NDM and OXA-181 carbapenemases in various countries, including Switzerland, Iraq, Canada, United Kingdom, India, and Italy [26, 46]. In the current study, two isolates of ST147 were detected which co-harboredblaNDM-1andblaOXA-48genes.
Carbapenem-resistance among K. pneumoniae isolates makes colistin the last therapeutic option for the treatment.
With the rise in consumption of colistin, cases of colistin resistant CRKP isolates are reported globally [1, 9, 11].
Chromosomal mutations inphoP/phoQ, pmrA/pmrB, mgrB and plasmid-borne mobile colistin resistance genes (mcr-1to mcr-9) positivity have an important role in increasing colistin resistance inK. pneumoniae[47, 10, 11]. The highest colistin resistance rate was reported in Asia (especially Korea and Singapore), followed by Europe (especially Greece) and America, where colistin resistance rates are continually increasing [48]. Nowadays, all known mcrgenes have been detected in variousK. pneumoniaeisolates, whereas a small number of studies have shown the presence ofmcrgenes in clinical K. pneumoniae isolates [49]. Several studies sug- gested that chromosomal mutations rather thanmcr genes positivity might have an important role in colistin resistance [47, 44, 50]. Different STs such as ST274, ST461, ST15, ST16, ST416, ST1890, ST37, ST1942, ST101, ST147, ST258, ST152, and ST15 were detected in carbapenem and colistin resistant K. pneumoniae [42, 50, 51]. We detected two ST types, ST101 (95.6%) and ST147 (4.4%) in carbapenem and colistin resistant K. pneumoniae. In Turkey, a few study investigatedmcr-1to-3genes among carbapenem and colistin resistant K. pneumoniae isolates and only Arabacı et al., determined mcr-1positive KRCP (5.2%) [44, 51–53]. In this study, we investigatedmcr-1 to-8genes in colistin and car- bapenem resistantK. pneumoniaeand reported the molecular characteristics ofmcr-1positive CRKP. To our knowledge, this is thefirst report ofmcr-1positiveK. pneumoniaeisolates that produce both NDM-1 and OXA-48 while also belonging to the one of the emerging clones ST147 from Turkey.
CONCLUSION
We identified two different STs, namely, ST101 and ST147 among the carbapenem and colistin resistantK. pneumoniae isolates identified during 2017 and 2019. ST101 is a epidemic ST and has been associated with OXA-48. Our results show that this ST also has the ability to develop colistin resistance. Early detection and surveillance can prevent the spread of carbapenem and colistin resistantK.
pneumoniaeisolates.
Ethical committee approval:Not required.
Conflict of interest:The authors declare no competing in- terests.
ACKNOWLEDGEMENTS
None.
SUPPLEMENTARY MATERIAL
Supplementary data to this article can be found online at https://doi.org/10.1556/030.2020.01275.
REFERENCES
1. Palmieri M, D'Andrea MM, Pelegrin AC, Mirande C, Brkic S, Cirkovic I, et al. Genomic epidemiology of Carbapenem- and Colistin-resistantKlebsiella pneumoniaeisolates from Serbia: pre- dominance of ST101 strains carrying a novel OXA-48 plasmid.
Front Microbiol 2020;11:294.
2. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of Carbapenemase-producingKlebsiella pneumoniae:
epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 2016;7:895.
3. Poirel L, Heritier C, Tol€un V, Nordmann P. Emergence of oxa- cillinase-mediated resistance to imipenem in Klebsiella pneumo- niae. Antimicrob Agents Chemother 2004;48:15–22.
4. Carr€er A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, et al.
Spread of OXA-48–encoding plasmid in Turkey and beyond.
Antimicrob Agents Chemother 2010;54:1369–73.
5. Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. Outbreak of OXA-48–positive carbapenem-resistantKlebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 2011;55:
2420–3.
6. Moquet O, Bouchiat C, Kinana A, Seck A, Arouna O, Bercion R, et al. Class D OXA-48 carbapenemase in multidrug-resistant enterobacteria, Senegal. Emerg Infect Dis 2011;17:143–4.
7. Benouda A, Touzani O, Khairallah MT, Araj GF, Matar GM. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann Trop Med Parasitol 2010;104:327–30.
8. Poirel L, Ros A, Carr€er A, Fortineau N, Carricajo A, Berthelot P, et al. Cross-border transmission of OXA-48–producing Enter- obacter cloacaefrom Morocco to France. J Antimicrob Chemother 2011;66:1181–2.
9. Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resis- tant and Carbapenemases-producing clinicalKlebsiella pneumoniae isolates. Infect Drug Resist 2019;12:3783–95.
10. Olaitan AO, Morand S, Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014;5:643.
11. Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance trends. Microbiol 2018;26:794–808.
12. Navon-Venezia S, Kondratyeva K, Carattoli A.Klebsiella pneumo- niae:a major worldwide source and shuttle for antibiotic resistance.
FEMS Microbiol Rev 2017;41:252–75.
13. EUCAST Clinical Breakpoints http://www.eucast.org/clinical_
breakpoints.
14. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010;65:490–5.
15. Lima Barbieri N, Nielsen DW, Wannemuehler Y, Cavender T, Hussein A, Yan SG, et al.mcr-1 identified in Avian Pathogenic Escherichia coli(APEC). PLoS One 2017;12:e0172997.
16. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, inEscherichia coli, Belgium, June 2016. Euro Surveill 2016;21.
17. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid- mediated colistin resistance gene mcr-3 inEscherichia coli. mBio 2017;8:e01166–17.
18. Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al.
Novel plasmid-mediated colistin resistance mcr-4 gene inSalmo- nellaandEscherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 2017;22.
19. Borowiak M, Fischer J, Hammerl JA, Hendriksen SR, Szabo I, Malorny B. Identification of a novel transposon-associated phos- phoethanolamine transferase gene,mcr-5, conferring colistin resis- tance in d-tartrate fermentingSalmonella enterica subsp. Enterica serovar Paratyphi B. J Antimicrob Chemother 2017;72:3317–24.
20. Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid- mediated colistin resistance gene mcr-7.1 inKlebsiella pneumoniae.
J Antimicrob Chemother 2018;73:1791–95.
21. Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 2018;7:122.
22. D’Agata EM, Gerrits MM, Tang YW, Samore M, Kusters JG. Com- parison of pulsed-field gel electrophoresis and amplified fragment- length polymorphism for epidemiological investigations of common nosocomial pathogens. Infect Control Hosp Epidemiol 2001;22:550–4.
23. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995;33:2233–9.
24. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005;43:4178–82.
25. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of di- versity. J Clin Microbiol 1988;26:2465–66.
26. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 2015;59:5873–84.
27. Meneks¸e S¸, Çag Y, Is¸ık ME, S¸ahin S, Hacıseyitoglu D, Can F, et al..
The effect of colistin resistance and other predictors on fatality among patients with bloodstream infections due toKlebsiella pneumoniaein an OXA-48 dominant region. Int J Infect Dis 2019;86:208–11.
28. Walther-Rasmussen J, Hoiby N. Class A carbapenemases. J Anti- microb Chemother 2007;60:470–82.
29. Deshpande LM, Rhomberg PR, Sader HS, Jones RN. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers:
report from the MYSTIC Program (1999–2005). Diagn Microbiol Infect Dis 2006;56:367–72.
30. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniaecarbapenemase-producing bacteria. Lancet Infect Dis 2009;9:228–36.
31. Labarca J, Poirel L, Ozdamar M, Turkogl€u S, Hakko E, Nordmann P. KPC-producingKlebsiella pneumoniae,finally targeting Turkey.
Labarca New Microbes New Infect 2014;2:50–1.
32. Tekeli A, Dolapci_I, Evren E, Oguzman E, Karahan ZC. Charac- terization ofKlebsiella pneumoniaecoproducing KPC and NDM-1 Carbapenemases from Turkey. Microb Drug Resist 2020;2:118–25.
33. Sagıroglu P, Hasdemir U, Altınkanat Gelmez G, Aksu B, Karatuna O, S€oyletir G. Performance of “RESIST-3 O.K.N. K-SeT” immu- nochromatographic assay for the detection of OXA-48 like, KPC, and NDM carbapenemases in Klebsiella pneumoniae in Turkey.
Braz J Mikrobiol 2018;4:885–90.
34. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 2015;59:5873–84.
35. Tekintas¸ Y, Çilli F, Eraç B, Yas¸ar M, Aydemir SS¸, Hos¸g€or Limoncu M. Comparison of phenotypic methods and polymerase chain re- action for the detection of carbapenemase production in clinical Klebsiella pneumoniaeisolates. Mikrobiyol Bul 2017;51:269–76.
36. Çakar A, Aky€on Y, G€ur D, Karatuna O,O€g€unç D,Ozhak Baysan B,€ et al. Investigation of carbapenemases in carbapenem-resistant Escherichia coli andKlebsiella pneumoniaestrains isolated in 2014 in Turkey. Mikrobiyol Bul 2016;1:21–33.
37. Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. European network on carbapenemases. Clin Microbiol Infect 2012;18:413–31.
38. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 2012;67:1597–606.
39. D€uzg€un A€O, Saral A. Next-generation sequencing of plasmid car- rying bla(OXA-48) in Klebsiella pneumoniae from Turkey. Acta Microbiol Immunol Hung 2019;66:261–72.
40. David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al.
Epidemic of carbapenem-resistantKlebsiella pneumoniaein Europe is driven by nosocomial spread. Nat Microbiol 2019;4:1919–29.
41. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase- producing enterobacteriaceae. Emerg Infect Dis 2011;17:1791–8.
42. Novovic K, Trudic A, Brkic S, Vasiljevic Z, Kojic M, Medic D, et al.
Molecular epidemiology of colistin resistant, carbapenemase-pro- ducing Klebsiella pneumoniaein Serbia from 2013 to 2016. Anti- microb Agents Chemother 2017;61:e02550–16.
43. Del Franco M, Paone L, Novati R, Giacomazzi CG, Bagattini M, Galotto C, et al. Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emer- gence of KPC-2 producingKlebsiella pneumoniaeclonal complex 101 (ST101 and ST1789). BMC Microbiol 2015;15:260.
44. Can F, Menekse S, Ispir P, Atac N, Albayrak O, Demir T, et al.
Impact of the ST101 clone on fatality among patients with colistin- resistantKlebsiella pneumoniaeinfection. J Antimicrob Chemother 2018;73:1235–41.
45. Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, et al. An update evolving epidemic ofblaKPC-2- carryingKlebsiella pneumoniaein Greece (2009–10). J Antimicrob Chemother 2011;66:1510–13.
46. Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveil- lance and molecular epidemiology ofKlebsiella pneumoniaeisolates that produce carbapenemases:first report of OXA-48-like enzyme- sin North America. Antimicrob Agents Chemother 2016;57:130–6.
47. Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P.
Resistance to colistin associated with a single amino acid change in protein PmrB amongKlebsiella pneumoniaeisolates of worldwide origin. Antimicrob Agents Chemother 2014;58:4762–6.
48. Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 2015;31:707–21.
49. Wang Y, Liu F, Hu Y, Zhang G, Zhu B, Gao GF. Detection of mobile colistin resistance gene mcr-9 in carbapenem-resistant Klebsiella pneumoniaestrains of human origin in Europe. J Infect 2020;80:578–606.
50. Longo LGA, de Sousa VS, Kraychete GB, Justo-da-Silva LH, Rocha JA, Superti SV, et al. Colistin resistance emerges in pandrug- resistantKlebsiella pneumoniaeepidemic clones in Rio de Janeiro, Brazil. Int J Antimicrob Agents 2019;54:579–86.
51. Guducuoglu H, Gursoy NC, Yakupogullari Y, Parlak M, Karasin G, Sunnetcioglu M, et al. Hospital outbreak of a colistin-resistant, NDM-1- and OXA-48-producingKlebsiella pneumoniae: high mor- tality from pandrug resistance. Microb Drug Resist 2018;24:966–72.
52. Borsa BA, Demirci M, Gungordu-Dalar Z, Karabiyik G, Aygun G, Kucukbasmaci O. Molecular mechanisms of colistin resistance amongKlebsiella pneumoniaestrains. Clin Lab 2019;65:1125–30.
53. ArabacıÇ, Dal T, Bas¸yigit T, Genis¸el N, Durmaz R. Investigation of carbapenemase and mcr-1 genes in carbapenem-resistantKlebsiella pneumoniaeisolates. J Infect Dev Ctries 2019;30:504–9.
Open Access. This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (https://
creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium for non-commercial purposes, provided the original author and source are credited, a link to the CC License is provided, and changes–if any–are indicated.