QUANTITATION OF DNA IN MONONUCLEAR PHAGOCYTES
D. O. Adams
I. PRINCIPLE
The specific activity of a given constituent within macro- phages can be expressed in relation either to content of pro- tein or to cell number· Determination of cell number is often advantageous, because cultivated macrophages may either produce large amounts of protein in culture, or lose protein after lysosomal discharge (1, 2 ) . Thus, the specific activity of a given constituent may rise or fall in the face of a falling or rising total amount of a particular enzyme. Determining cell number by hemacytometer count is often inaccurate in this cir- cumstance, because a large but variable number of macrophages may be lysed while removing them from the culture vessel (3).
Determination of DNA content provides an accurate, reliable, and sensitive method for quantifying cell number (6). On the other hand, determination of specific activity in regard to cellular protein does take into account variations in the size of different macrophages.
METHODS FOR STUDYING Copyright © 1981 by Academic Press, Inc.
MONONUCLEAR PHAGOCYTES 3 3 1 All rights of reproduction in any form reserved.
ISBN 0-12-044220-5
The present assay, devised specifically for quantifying DNA in mononuclear phagocytes, offers the advantages of sim- plicity and sensitivity (4). The DNA of macrophages lysed in detergent is precipitated by cold PCA, extracted in hot PCA, and quantified by spectrophotometry after reaction with di- phenylamine (5). The assay can be applied to macrophages in suspension or adherent to culture vessels such as petri dishes or 16 mm wells. As few as 1 x 10^ macrophages can be quanti-
fied by this procedure, and the assay is linear over the range 1 - 50 yg of DNA/sample.
II. REAGENTS
PCA. 7 M Perchloric Acid (PCA) and 1 M PCA
Diphenylamine: 3 gm % Diphenylamine (Eastman Kodak, Rochester, New York). Under a fume hood, dissolve 3 mg/100 ml glacial acetic acid. Store under a fume hood in an amber
bottle at room temperature. This reagent is generally good for 2 - 3 months.
Acetaldehyde: 1.6 gm % Acetaldehyde (Eastman Kodak). On ice under a fume hood, add 0.5 ml of acetaldehyde stock to 24.5 ml of distilled water. Store in an amber bottle with a screw top at 4°C. This is usually good to 2 - 3 months but often deteriorates to acetic acid. If the assay begins to give erratic results, check the acetaldehyde first.
DNA Standard. Dissolve 8 mg of DNA (Sigma No. D-1501;
Type 1, Na salt) in 20 ml of 5 mM NaOH. When thoroughly dis- solved, make a 1 : 5 dilution in 5 mAf NaOH. Determine A260nm and A280nm· T^e D N A content of the dilution in microgram per milliliter is then calculated using the nomogram of Warburg and Christian (Fig. 1) (6). Store the stock at 4°C, where it is good for about 6 months.
III. PROCEDURE
A, Cell Preparation
(1) It is important to keep the cells at 4°C at all times.
(2) Method 1: Wash cultured cells in wells or plates and then freeze at -70°C in an appropriate volume of PBS (i.e., 0.5 ml PBS/16 m m w e l l ) . Thaw on ice. To each w e l lf add 0.5 ml of freshly prepared 0.4% Triton X-100. Incubate on ice
y 2 0 0 0
4 1.900 T 1800 4- I 700 4 1600 4 · 500 -1-1400 4- 1.300 4-1200 4 1100 4 1.000 -I- 0.900 4 0B00 4 0.700 4 0.600 4 0.500 4 0400 4 0300 4 0200 4 0J00
-*- 0O00280
3
26
[-2000 M 900
11800
H.700 M 6 0 0
11500
M.400 H 300 j- 1.200 Γ 1.100
L looo
j. 0.900
1 O800 - 0 7 0 0
■ 0.600
• 0.500 - 0 4 0 0
" 0.300 - 0 2 0 0 - 0100 - 0 0 0 0
>0
j 056
4 054 4 052 4-050 + 048 4- 046 4" .044 4" .042 + 040 + .038 4-.036 4-034 4- .032 4- .030 4" .028 4-.026 4-024 4 022 + .020 4 .018 4 .016 4 .014 4 .012 4 .010 4 008 4 .006 4 .004 4 .002 -*- 000
DNA
Fig. 1. Determination of DNA by A260
an<^
Ά280 (6). Using a cuvette with a 1-cm light path, determine A2S0nm
and A280nm·
From these, the concentration of DNA (milligram per liter) can
be determined. Based on a nomogram prepared by E. Adams from
Calbiochem Corp. for biochemical research. (With permission
of Calbiochem Corp.)
for 30 min. Scrape up cells with a rubber policeman. Trans- fer samples completely to test tubes on ice.
(3) Method 2: To cells at 4°C in a culture dish, add 0.4%
Triton X-100 to a final concentration of 0.2%. Incubate 30 min at 4°C. Remove by scraping. Freeze in a Dry Ice - acetone slurry. Transfer samples completely to and store in polypro- pylene tubes at -70°C. Thaw on ice.
B. Determination of DNA
(1) Samples and standards are assayed in triplicate.
(2) Standards are made from DNA stock in 5 mM NaOH; 14, 7, 3.5, 1.75, and 0.87 yg of DNA/ml provide a useful range.
Place 1.0 ml of each standard in 10 x 75 disposable culture tubes. Prepare blanks of 1.0 ml of NaOH and PBS. Add 20 y£
of 10% Triton X-100 to each standard and incubate on ice 30 min. Add 30 \iZ of 7 M PCA to each standard. Mix thoroughly.
(3) To 10 x 75 mm disposable tubes on ice containing 30 ]ii 1 M PCA, add 1.0 ml of each lysate sample. Mix thoroughly on a vortex mixer.
(4) The DNA should now be stable. However, keep samples on ice. From this point on, all standards and unknown are identically treated.
(5) Incubate PCA-treated samples on ice for 30 min. Spin samples at 10,000 g for 15 min at 4°C. The DNA is now pelleted.
(6) Gently aspirate the supernatant with a Pasteur pipette while continuously observing the residual pellet. Discard supernatant carefully.
(7) Add 0.5 ml of hot PCA (70°C) and incubate 30 min at 70°C after covering the tubes with parafilm to hydrolyze the residual DNA.
(8) Remove the hydrolysate containing the DNA completely from each tube and place it in a clean 10 x 75 mm glass tube.
(9) Prepare the colorimetric reagent just before use:
10 ml of 3% diphenylamine, 0.2 ml of cone. H2S04, 0.1 ml of 1.6% acetaldehyde.
(10) Add 0.5 ml of the above reagent to each sample, blank or standard. Seal tubes with parafilm. Incubate 16-24 hr at 37°C. The time can be varied as long as all samples are simi- larly treated.
(11) Spin at 10,000 g for 10 min at 4°C.
(12) Determine AçQOnm against a water blank.
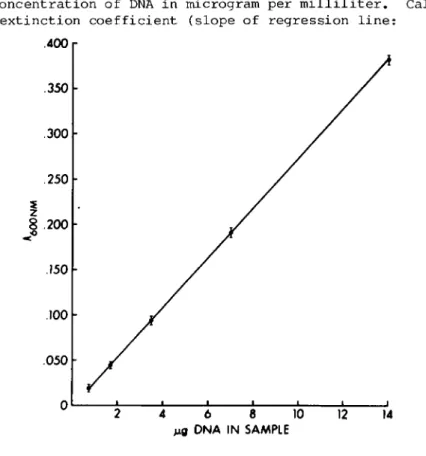
IV. CALCULATION OF DATA
P l o t n e t A^QQnm of s t a n d a r d s ( v e r s u s NaOH b l a n k ) a g a i n s t t h e c o n c e n t r a t i o n of DNA i n m i c r o g r a m p e r m i 1 1 i l i t e r . C a l c u - l a t e e x t i n c t i o n c o e f f i c i e n t ( s l o p e of r e g r e s s i o n l i n e :
.400
.350
.300
.250
8 .200
.150
.100 .050
2 4 6 8 10 12 14
Mg DNA IN SAMPLE
Fig. 2. Ά standard curve for DNA.
Net AçQOnm/MÇJ DNA) . A typical standard curve is shown in Fig. 2. To calculate DNA content of unknowns falling on the linear part of the graph, divide the net A^oOnm (versus the PBS blank) by the extinction coefficient.
J I I I L
V . COMMENTS
The assay is sensitive to 1 yg DNA or 100,000 macrophages (4). 1 x 10^ murine peritoneal macrophages contain
10.1 ± 0 . 3 6 g DNA (4). The coefficient of variation between triplicate samples should be approximately ±2%. The principal problems with assaying small amounts of DNA are in complete precipitation of DNA (Section III. B, step 5 ) , avoidance of loss of DNA (Section III. B, step 6 ) , and complete recovery of the DNA (Section III. B, step 8 ) . The reproducibility of the assay can be increased by using samples containing the lysate of ^ 5.0 x 105 macrophages, but this should not be necessary if the assay is carefully performed.
Acknowledgment
Supported in part by USPHS Grant 16784.
REFERENCES
Z. A. Cohn and B. Benson. The differentiation of mono- nuclear phagocytes: Morphology, cytochemistry, and bio- chemistry. J. Exp. Med. 121: 153-169, 1965.
J. Schnieder and M. Baggiolini. Secretion of lysosomal hydrolases by stimulated and nonstimulated macrophages.
J. Exp. Med. 148: 435-450, 1978.
D. 0. Adams. Macrophages. In "Methods in Enzymology,"
Vol. LVIIIf Cell Culture (W. Jakoby and I. Pastan, eds.), pp. 494-505. Academic Press, New York, 1979.
S. L. Cookson and D. O. Adams. A simple, sensitive assay for determining DNA in mononuclear phagocytes and other leukocytes. J. Immunol. Methods 23: 169-173, 1978.
K. Burton. A study of the conditions and mechanisms of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem. J. 62: 315, 1956.
0. Warburg and W. Christian. Isolierung und Kristallisa- tion des Garungsferments Enolase. Biochem. Z. 310: 384- 421, 1940.