Acta Microbiologica et Immunologica Hungarica
DOI:10.1556/030.66.2019.040
© 2019 Akadémiai Kiad´o, Budapest
ORIGINAL ARTICLE
* Corresponding author:
Amira ElBaradei
Department of Microbiology and Immunology, Faculty of Pharmacy and Drug Manufacturing, Pharos University in Alexandria, Canal El Mahmoudia Street, beside Green Plaza Complex, Alexandria, Egypt;
Phone: +20 3 3877032/033; +20 01 205233907; Fax: +20 3 3877149;
E-mail:amiraelbaradei@gmail.com
parC mutations, and AbaQ ef fl ux pump among Acinetobacter baumannii clinical isolates causing ventilator-associated pneumonia
NANCY M. ATTIA
1and AMIRA ELBARADEI
2,3*
1Microbiology Department, Medical Research Institute, Alexandria University, Alexandria, Egypt
2Department of Microbiology and Immunology, Faculty of Pharmacy and Drug Manufacturing, Pharos University in Alexandria, Alexandria, Egypt
3Alexandria University Hospital, Alexandria University, Alexandria, Egypt
Received: September 21, 2019•Accepted: October 14, 2019
ABSTRACT
Acinetobacter baumannii has emerged as an important nosocomial pathogen due to its ability to survive in hospital settings and its antimicrobial resistance. It is one of the key pathogens in ventilator- associated pneumonia (VAP). The aim of this study was to characterize the mechanisms of quinolone resistance amongA. baumanniiisolates causing VAP and to investigate the presence of the novelabaQ gene among them. Quinolone-resistant A. baumannii isolates causing VAP were collected over a period of 4 months. Mutations withingyrAandparCwere analyzed and the presence ofqnrA,qnrB, qnrS, and abaQ was investigated genotypically. Twenty-one A. baumanniiisolates were collected, most of them (76.2%) were extensively drug-resistant (XDR) and only one isolate (4.8%) was pandrug-resistant (PDR). All isolates showed high level of resistance to ciprofloxacin, while qnrA, qnrBandqnrSwere absent among our isolates. This is thefirst report ofA. baumanniiisolates co- harboring Ser81Leu ingyrAand Ser84Leu inparCtogether with the novelabaQgene. Interestingly, a new mutation ingyrAquinolone resistance-determining region Arg89Cys was detected among two of our isolates. The emergence of XDR and PDR isolates among A. baumannii causing VAP is an alarming threat.
KEYWORDS
quinolone resistance,gyrA,parC,abaQ,Acinetobacter baumannii, MALDI-TOF MS
INTRODUCTION
Acinetobacter baumannii, is a Gram-negative, aerobic, non-lactose fermenting bacteria, which was deemed a low-category pathogen. However, it has emerged as a significant nosocomial pathogen, causing bloodstream, respiratory tract, and urinary tract infections [1]. In addition, A. baumannii has become a momentous cause of ventilator-associated pneumonia (VAP) worldwide, which is a frequent nosocomial infection among critically ill patients to which high rates of morbidity and mortality have been linked [1, 2].
More attention has been drawn to A. baumannii due to its growing antimicrobial resistance. A. baumannii shows intrinsic resistance to different antibacterial agents such as aminopenicillins, cephalosporins of thefirst and second generation, and chloramphenicol [3].
According to the 2016 guidelines of the American Thoracic Society and Infectious Diseases Society of America (ATS-IDSA) [4], quinolones are among the proposed empiric options for clinically suspected VAP. However, quinolones as well as other antibiotics are increasingly 67 (2020) 4, 234–238
rendered inactive due to various acquired resistance mechan- isms by A. baumannii [5]. Hence, A. baumannii causing VAP are often therapeutically challenging and are therefore associated with high mortality [6].
Quinolone resistance has progressively spread worldwide.
Resistance to quinolones is mainly mediated by mutations in the quinolone resistance-determining region (QRDR) of the DNA gyrase gene (gyrA) and/or topoisomerase IV gene (parC). These substitutions are primarily focused in the amino terminal domains of GyrA and ParC, which refer to residues 67 to 106 and residues 63 to 102 located inEscher- ichia coli, respectively [7].
Another mechanism for quinolone resistance is the protection of the DNA by the inhibition of quinolone binding to DNA gyrase and topoisomerase. This is mediated via plasmid-mediated quinolone-resistance (PMQR) deter- minants, which includeqnrA, qnrB, and qnrS[8].
Recently, a novel efflux pump has been described inA.
baumannii. It is a putative major facilitator superfamily (MFS) transporter, which is encoded by abaQ gene.
This efflux pump is the first to mediate quinolone resis- tance amongA. baumanniiisolates. Moreover, it plays an important role in surface-associated motility and in virulence [9].
Our aim was to characterize the mechanisms of quinolone resistance and to investigate the presence of the novel abaQ gene among A. baumannii clinical isolates causing VAP.
MATERIALS AND METHODS Strains
A. baumanniiisolates resistant to both ciprofloxacin (CIP) and levofloxacin (LEV) were collected from clinical samples of VAP submitted to the Microbiology Department, Medical Research Institute, Alexandria University. These isolates were collected and included in this study over a period of 4 months (starting from September 2016 to the end of December 2016).
The identification of these isolates as A. baumannii was confirmed using matrix-assisted laser desorption ionization– time offlight mass spectrometry (MALDI-TOF MS; Bruker, Billerica, MA, USA) as well as the presence ofblaOXA-51gene.
Antimicrobial susceptibility testing
Disk diffusion method was used for the antimicrobial sus- ceptibility testing of the collected strains. All antibiotic disks were chosen according to the CLSI recommendations [10]; these were ampicillin/sulbactam, aztreonam (TZP), ceftazidime, cefepime (FEP), cefotaxime (CTX), ceftriaxone (CRO), imipenem (IPM), meropenem (MEM), colistin (CT), gentamicin, tobramycin, amikacin (AK), doxycycline (DO), CIP, LEV, and trimethoprim/sulfamethoxazole. The strains were reported as susceptible, intermediate, or resistant to the previously mentioned agents according to the CLSI M100-S28 breakpoints [10]. These breakpoints were also Table I.Primers used in this study
Primer Nucleotide sequence (5′–3′) Amplicon length in bases Reference
blaOXA-51(F) TAA TGC TTT GAT CGG CCT TG 350 [11]
blaOXA-51(R) TGG ATT GCA CTT CAT CTT GG
gyrA(F) CGACCGATTGCCATTGAGGA 682 This study
gyrA(R) CGGTACGGTAGGCATCAACA
parC(F) CAGAAAACCGCTCTGTAGCC 862 This study
parC(R) TCATGATCCGATTCATCACG
qnrA(F) AGAGGATTTCTCACGCCAGG 661 [12]
qnrA(R) TGCCAGGCACAGATCTTGAC
qnrB(F) GGMATHGAAATTCGCCACTG 562 [12]
qnrB(R) TTTGCYGYYCGCCAGTCGAA
qnrS(F) GCAAGTTCATTGAACAGGGT 605 [12]
qnrS(R) TCTAAACCGTCGAGTTCGGCG
abaQ(F) GCTGCCAACTGCATAACTGG 490 This study
abaQ(R) GCTGGCAATGGTTGTTCGTT
Note:All primers were purchased from Invitrogen by Thermo Fisher Scientific (CA, USA); the PCR Master mix used was MyTaq Red Mix, which was obtained from BioLine (London, UK). F: forward; R: reverse.
used to determine CIP and LEV MIC values, which were obtained using broth microdilution method [10].
Detection of resistance genes
Genotypic detection of qnrA, qnrB, qnrS, and the newly described efflux pumpabaQwas performed using polymerase chain reaction (PCR). Detection ofgyrAandparCmutations was carried out using PCR followed by sequencing of the amplified products using the chain-termination method. Muta- tions within the QRDR were determined by BioEdit sequence alignment editor, using the control sequence for gyrA (NZ_CZWC01000014.1) and parC (NZ_CZWC01000100.1) inA. baumannii. Then, alignments using Clustal Omega were applied to determine the position of these mutations in relation toE. coli gyrAandparCsequences. Primers, used in this study, are listed in (TableI). Some of these primers were designed in this study using Primer3 (http://www.ncbi.nlm.nih.gov/tools/
primer-blast/).
RESULTS
Twenty-one A. baumanniiisolates were included in this study. These isolates were collected from different samples obtained from VAP including endotracheal tubes, sputum,
and bronchoalveolar lavage samples. Their identification asA. baumannii isolates was confirmed by MALDI-TOF MS as well as the presence ofblaOXA-51gene among all of them.
All the isolates resistant to both CIP and LEV were also resistant to TZP, FEP, CTX, CRO, IPM, MEM, and AK. Only one isolate was found CT-resistant. The disk diffusion test results are listed in TableII.
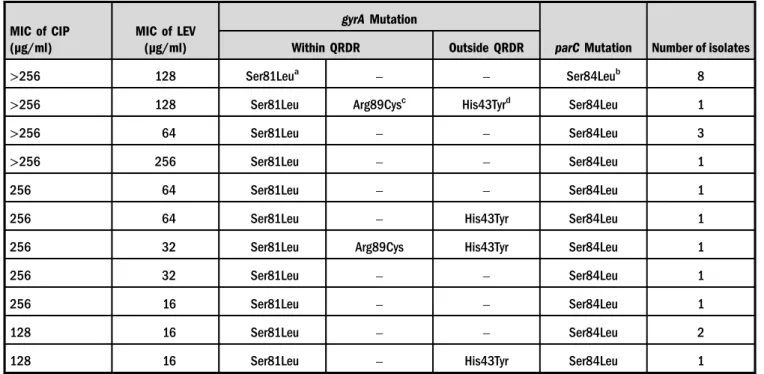
MIC values of CIP revealed that all of the isolates showed high-level resistance to CIP (≥32 μg/ml), whereas MIC values of LEV ranged from 16 to 256μg/ml. Details of the MIC and the detected mutations ingyrAandparCvalues are shown in TableIII.
Genotypically, abaQgene was detected among all of our isolates, whereasqnrA, qnrB, andqnrS were absent.
DISCUSSION
VAP is considered one of the most commonly acquired nosocomial infections. It is among the most significant nosocomial infections caused byA. baumannii [6]. More- over, multidrug-resistant (MDR), extensively drug-resistant (XDR), and even pandrug-resistant (PDR) A. baumannii isolates are continuously being reported [5,13]. Among our Table II.Susceptibility patterns of the 21A. baumanniiisolates
Antibiotic
Resistance Intermediate Sensitive
N % N % N %
Ampicillin/sulbactam 20 95.24 1 4.76 0 0
Piperacillin/tazobactam 21 100 0 0 0 0
Ceftazidime 20 95.24 0 0 1 4.76
Cefepime 21 100 0 0 0 0
Cefotaxime 21 100 0 0 0 0
Ceftriaxone 21 100 0 0 0 0
Imipenem 21 100 0 0 0 0
Meropenem 21 100 0 0 0 0
Colistin 1 4.76 0 0 20 95.24
Gentamicin 19 90.48 1 4.76 1 4.76
Tobramycin 19 90.48 0 0 2 9.52
Amikacin 21 100 0 0 0 0
Doxycycline 6 28.57 5 23.81 10 47.62
Ciprofloxacin 21 100 0 0 0 0
Levofloxacin 21 100 0 0 0 0
Trimethoprim/sulfamethoxazole 18 85.72 2 9.52 1 4.76
21A. baumanniiisolates, 1 (4.8%) was PDR, 4 (19%) were MDR, whereas the rest of the isolates (76.2%) were XDR.
Huang et al. [6] reported that all the isolates in his retro- spective study were MDR. According to Ciginskien et al. [14], the percentages of MDR and XDR A. baumannii were (13.3%) and (68.3%), respectively. The continuing occur- rence of XDR and PDRA. baumanniiisolates from VAP is an alarming threat.
All the isolates in this study harbored mutations within the QRDR ofgyrAandparC. The primary mutation (Ser81- Leu in gyrA) was present in all the isolates as well as the secondary mutation (Ser84Leu inparC). Interestingly, both mutations were found together in 100% of our isolates.
According to Ostrer et al. [15], these two mutations are enough to predict CIP and LEV resistance. Furthermore, a new mutation was detected ingyrAQRDR in the position 89 (Arg89Cys), which is equivalent to position 91 inE. coli gyrA.
This substitution resulted in a polar non-charged amino acid (cysteine) instead of the basic amino acid (arginine). Akter et al. [16] reported the critical role of arginine at the position 91 ofE. coli gyrA,where it stabilizes the drug-protein putative binding pocket through H-bonding interactions with CIP and another interaction with Asp at position 87 [16]. To the best of our knowledge, this mutation has not been reported before inA. baumannii. However, a silent mutation in this position has been reported previously in E. coli by Conrad et al. [17] and Lehn et al. [18].
In this study, qnrA, qnrB, and qnrS were not detected among our isolates. Several studies found these genes to be absent among theirA. baumanniiisolates [19,20]. However, Hamed et al. [21] found only one strain-harboringqnrSout of his 28A. baumanniiisolates, with the absence ofqnrAand qnrBgenes.
On the other hand,abaQgene was found among all of our isolates. It encodes the AbaQ efflux pump, which is involved in both pathogenicity and quinolone resistance [9].
Perez-Varela et al. [9] reported that abaQ was generally present amongA. baumanii clinical isolates. However, this is the first study to investigate its presence among A. baumanniicausing VAP in Egypt.
This study highlights the mechanisms of quinolone resistance amongA. baumannii causing VAP. Mutations within QRDR are the main mechanism underlying quinolone resistance among our isolates. This is thefirst study to report a new mutation in the QRDR of A.
baumannii gyrA (in position 89). This mutation needs further elucidation. On the other hand, qnrA, qnrB, and qnrS genes had no effect on quinolone resistance among our isolates. The significance of the new AbaQ efflux pump encoding gene needs further investigations. The increased XDR A. baumannii isolated from VAP repre- sents a huge obstacle to achieving prompt appropriate antimicrobial therapy, which is crucial to enhance clinical outcomes.
Table III.MIC values of ciprofloxacin (CIP) and levofloxacin (LEV) and amino acid substitutions withingyrAandparCgenes for all isolates
MIC of CIP (μg/ml)
MIC of LEV (μg/ml)
gyrAMutation
parCMutation Number of isolates
Within QRDR Outside QRDR
>256 128 Ser81Leua – – Ser84Leub 8
>256 128 Ser81Leu Arg89Cysc His43Tyrd Ser84Leu 1
>256 64 Ser81Leu – – Ser84Leu 3
>256 256 Ser81Leu – – Ser84Leu 1
256 64 Ser81Leu – – Ser84Leu 1
256 64 Ser81Leu – His43Tyr Ser84Leu 1
256 32 Ser81Leu Arg89Cys His43Tyr Ser84Leu 1
256 32 Ser81Leu – – Ser84Leu 1
256 16 Ser81Leu – – Ser84Leu 1
128 16 Ser81Leu – – Ser84Leu 2
128 16 Ser81Leu – His43Tyr Ser84Leu 1
Note:MIC: minimum inhibitory concentration; QRDR: quinolone resistance-determining region.
aEquivalent to Ser83Leu inE. coli gyrA.
bEquivalent to Ser80Leu inE. coli parC.
cEquivalent to Arg91Cys inE. coli gyrA.
dEquivalent to His45Tyr inE. coli gyrA.
Acknowledgements:This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
1. Deccache, Y., Irenge, L. M., Savov, E., Ariciuc, M., Macovei, A., Trifonova, A., Gergova, I., Ambroise, J., Vanhoof, R., Gala, J. L.:
Development of a pyrosequencing assay for rapid assessment of quinolone resistance inAcinetobacter baumanniiisolates. J Microbiol Methods86, 115–118 (2011).
2. Erdem, H., Cag, Y., Gencer, S., Uysal, S., Karakurt, Z., Harman, R., Aslan, E., Mutlu-Yilmaz, E., Karabay, O., Uygun, Y., Ulug, M., Tosun, S., Dogru, A., Sener, A., Dogan, M., Hasbun, R., Durmus, G., Turan, H., Batirel, A., Duygu, F., Inan, A., Akkoyunlu, Y., Celebi, G., Ersoz, G., Guven, T., Dagli, O., Guler, S., Meric-Koc, M., Oncu, S., Rello, J.: Treatment of ventilator-associated pneumonia (VAP) caused byAcinetobac- ter: Results of prospective and multicenter ID-IRI study. Eur J Clin Microbiol Infect Dis (2019).
3. Cherkaoui, A., Emonet, S., Renzi, G., Schrenzel, J.: Character- istics of multidrug-resistant Acinetobacter baumanniistrains isolated in Geneva during colonization or infection. Ann Clin Microbiol Antimicrob14, 42 (2015).
4. Kalil, A. C., Metersky, M. L., Klompas, M., Muscedere, J., Sweeney, D. A., Palmer, L. B., Napolitano, L. M., O’Grady, N. P., Bartlett, J. G., Carratala, J., El Solh, A. A., Ewig, S., Fey, P. D., File, T. M., Jr., Restrepo, M. I., Roberts, J. A., Waterer, G. W., Cruse, P., Knight, S. L., Brozek, J. L.: Management of adults with hospital-acquired and ventilator-associated pneu- monia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis63, e61–e111 (2016).
5. Nowak, J., Zander, E., Stefanik, D., Higgins, P. G., Roca, I., Vila, J., McConnell, M. J., Cisneros, J. M., Seifert, H., MagicBullet Working Group, W. P.: High incidence of pandrug-resistant Acinetobacter baumanniiisolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother 72, 3277–3282 (2017).
6. Huang, Y., Zhou, Q., Wang, W., Huang, Q., Liao, J., Li, J., Long, L., Ju, T., Zhang, Q., Wang, H., Xu, H., Tu, M.:Acinetobacter baumanniiventilator-associated pneumonia: Clinical efficacy of combined antimicrobial therapy andin vitrodrug sensitivity test results. Front Pharmacol10, 92 (2019).
7. Hooper, D. C., Jacoby, G. A.: Mechanisms of drug resistance:
Quinolone resistance. Ann N Y Acad Sci1354, 12–31 (2015).
8. Asif, M., Alvi, I. A., Rehman, S. U.: Insight intoAcinetobacter baumannii: Pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist11, 1249–1260 (2018).
9. Perez-Varela, M., Corral, J., Aranda, J., Barbe, J.: Functional Characterization of AbaQ, a novel efflux pump mediating
quinolone resistance inAcinetobacter baumannii. Antimicrob Agents Chemother62, e00906–e00918 (2018).
10. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.
CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA, 2018.
11. Turton, J. F., Woodford, N., Glover, J., Yarde, S., Kaufmann, M. E., Pitt, T. L.: Identification ofAcinetobacter baumanniiby detection of theblaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol44, 2974–2976 (2006).
12. Srinivasan, V. B., Rajamohan, G., Pancholi, P., Stevenson, K., Tadesse, D., Patchanee, P., Marcon, M., Gebreyes, W. A.:
Genetic relatedness and molecular characterization of multi- drug resistant Acinetobacter baumannii isolated in central Ohio, USA. Ann Clin Microbiol Antimicrob8, 21 (2009).
13. Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., Monnet, D. L.: Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect18, 268–281 (2012).
14. Ciginskiene, A., Dambrauskiene, A., Rello, J., Adukauskiene, D.: Ventilator-associated pneumonia due to drug-resistant Acinetobacter baumannii: Risk factors and mortality relation with resistance profiles, and independent predictors of in- hospital mortality. Medicina (Kaunas)55, E49 (2019).
15. Ostrer, L., Khodursky, R. F., Johnson, J. R., Hiasa, H., Khodursky, A.: Analysis of mutational patterns in quinolone resistance-determining regions of GyrA and ParC of clinical isolates. Int J Antimicrob Agents53, 318–324 (2019).
16. Akter, F., Amin, M. R., Osman, K. T., Anwar, M. N., Karim, M. M., Hossain, M. A.: Ciprofloxacin-resistantEscherichia coliin hospital wastewater of Bangladesh and prediction of its mechanism of resistance. World J Microbiol Biotechnol28, 827–834 (2012).
17. Conrad, S., Oethinger, M., Kaifel, K., Klotz, G., Marre, R., Kern, W. V.:gyrAMutations in high-levelfluoroquinolone-resistant clinical isolates ofEscherichia coli. J Antimicrob Chemother38, 443–455 (1996).
18. Lehn, N., Stower-Hoffmann, J., Kott, T., Strassner, C., Wagner, H., Kronke, M., Schneider-Brachert, W.: Characterization of clinical isolates ofEscherichia colishowing high levels offluoro- quinolone resistance. J Clin Microbiol34, 597–602 (1996).
19. Higuchi, S., Shikata, M., Chiba, M., Hoshino, K., Gotoh, N.:
Characteristics of antibiotic resistance and sequence type of Acinetobacter baumanniiclinical isolates in Japan and the antibac- terial activity of DS-8587. J Infect Chemother20, 256–261 (2014).
20. Bakour, S., Touati, A., Sahli, F., Ameur, A. A., Haouchine, D., Rolain, J. M.: Antibiotic resistance determinants of multidrug- resistantAcinetobacter baumanniiclinical isolates in Algeria.
Diagn Microbiol Infect Dis76, 529–531 (2013).
21. Hamed, S. M., Elkhatib, W. F., El-Mahallawy, H. A., Helmy, M. M., Ashour, M. S., Aboshanab, K. M. A.: Multiple mechan- isms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci Rep 8, 12268 (2018).