ContentslistsavailableatScienceDirect
International Journal of Biological Macromolecules
jo u r n al hom e p ag e :w w w . e l s e v i e r . c o m / l o c a t e / i j b i o m a c
Helicoverpa-inducible Thioredoxin h from Cicer arietinum: structural modeling and potential targets
Archana Singh
a,1, Chetna Tyagi
b,1, Onkar Nath
c, Indrakant K. Singh
c,d,∗aDepartmentofBotany,HansRajCollege,UniversityofDelhi,Delhi,110007,India
bDepartmentofMicrobiology,UniversityofSzeged,Középfasor52.,6726,Szeged,Hungary
cMolecularBiologyResearchLaboratory,DeshbandhuCollege,UniversityofDelhi,Kalkaji,NewDelhi,110019,India
dDepartmentofEntomology,UniversityofKentucky,S-225AG.Science,North,Lexington,KY40546-0001,UnitedStates
a r t i c l e i n f o
Articlehistory:
Received1September2017
Receivedinrevisedform9December2017 Accepted12December2017
Availableonline17December2017
Keywords:
Herbivory Chickpea Thioredoxinh Redoxregulation Comparativemodeling Docking
a b s t r a c t
Thioredoxinsaresmallanduniversalproteins,whichareinvolvedinthecellredoxregulation.Inplants, theyparticipateinabroadrangeofbiochemicalprocesseslikeself-incompatibility,seedgermination, pathogen&pestdefenseandoxidativestresstolerance.Theh-typeofthioredoxin(Trx-h)proteinrepre- sentsthelargestTrxfamily.Herein,wecharacterizedtheHelicoverpa–inducibleTrxhfromanimportant legume,Cicerarietinum,CaHaTrx-h,‘CGFS’typeTrxs,whichencodesfora113aminoacidslongprotein andpossesscharacteristicmotifs“FLKVDVDE”and“VVDFTASWCGPCRFIAPIL”and73%sequenceidentity withAtTrx-h.Homologymodelingandsimulationofthetargetshowedthattheextendedß-sheetregions remainstableduringthesimulationwhilethehelicalregionsfluctuatebetweenalphaand3-10helical formsandhighlightstheflexibilityofhelix2-helix3andterminalregionsprobablytoaccommodatean approachingproteintargetandfacilitatetheirinteraction.Duringthesimulation,thestructureexists infiveenergyminimaclusterswithbiggestclustersizebelongingto20–25nstimeframes.PR-5and MannitolDehydrogenasewerenominatedaspotentialtargetsandsharecloseinteractionwithCaHaTrx- hviadisulfidebondreduction.Thestudyisaneffortinthedirectionofunderstandingstress-related mechanismsincropplantstoovercomelossesinagriculturalyield.
©2017ElsevierB.V.Allrightsreserved.
1. Introduction
Thioredoxins (Trxs)are a class of small disulfidereductases thatregulatevariousredox-dependentprocesses[1–3].Trxswere discoveredin1964in anE.colisystemasanelectron donorof ribonucleotide reductase (anenzyme involved in DNA synthe- sis) that ledtoa great dealof interest in itsmultifariousroles [4].Theyhavemultiplesub-cellularlocalizationssuchascytosol, nucleus,mitochondria,andchloroplastortheymaybereleasedto theextracellularenvironment[5,6].Outofsixmajorcategoriesof ArabidopsisTrxconstitutingh,f,m,o,x,andybasedondifferent sub-cellularlocalization,Trx-hrepresentsthelargeststructurally andbiochemicallycomplicatedfamily[2,7].Trxsplayasignificant roleinthemaintenanceofcellularredoxhomeostasisasreduc- tases[7–9], regulateprogrammedcelldeathviadenitrosylation,
∗ Correspondingauthorat:MolecularBiologyResearchLaboratory,Department ofZoology,DeshbandhuCollege,UniversityofDelhi,Kalkaji,NewDelhi,110019, India.
E-mailaddress:iksingh@db.du.ac.in(I.K.Singh).
1 Theseauthorscontributedequally.
andalsoaidindefenseresponses[10–12].Thetoxicityofreactive oxygenspecies(ROS)suchashydrogenperoxideissequesteredby Trx-hproteinsthroughathiol-disulfideexchangemechanism[13].
Anactivesitedithiol,presentinCXXCmotif,partiallyexposed atthetargetproteinsurfaceisthekeyplayerinbreakingofdisul- fidebondsinoxidizedsubstrateproteins.Thesecysteineresidues areoxidizedandformadisulfide,whichisconvertedbacktothe reducedformbyNADPH-dependentthioredoxinreductase.There seemstobeaminorbutcrucialstructuraldifferencebetweenthe reducedandoxidizedformsofTrxandismainlytodo withthe surfaceelectronegativityofthedithiolbasedonitsredoxpotential [14,15].Inaddition,anexampleoftheirroleindefenseresponse hasbeen evidenced in Arabidopsis and it hasbeen shown that the expressionof AtTrx-h3and AtTrx-h5is induced bycertain pathogens and theycontribute to systemic acquired resistance (SAR)[10,16].WefocusedourinvestigationonTrx-hproteinfrom Cicerarietinumtorealizeitsfunctioninplantdefense.
Chickpea (Cicer arietinum) is an important food cropin the middle-easternandsoutheasternpartsoftheworldincludingIndia.
Helicoverpaarmigerainfestationhasbeenamajorconstraintfor successfulproductionofchickpea[17,18].Inourpreviousstudy, https://doi.org/10.1016/j.ijbiomac.2017.12.079
0141-8130/©2017ElsevierB.V.Allrightsreserved.
232 A.Singhetal./InternationalJournalofBiologicalMacromolecules109(2018)231–243
wehaveshownthatTrxhomologofchickpeanamely,CaHaTrx-h changesitstranscriptlevelduringherbivoryandtreatmentwith signalingcompounds.Here,wereportfurthercharacterizationof CaHaTrx-h.Thefull-lengthsequenceofCaHaTrx-hwasretrieved andacomputationalstudytoelucidateitsevolutionaryrelation- shipswithothersimilarproteinswasconducted.Itisalsocrucial to understandits expression mechanism and to know the co- expressedgenesandtheirregulation.Ashortmoleculardynamics based simulation can also give majorinsight into dynamics of its structure, which is otherwise, wellknown for other model specieslikeArabidopsis.Toconfirmthatitinteractandregulates otherproteins,twowell-knowntargetsofAtTrx-hwereselected.
Thesetargetshavebeenknowntobeimplicatedinplantdefense responses and thus, can be correlated with our experimental results. This study throws light on the structural details, sub- cellularlocation,andinteractiondynamicsofCaHa-Trx-h.Theuse oftimeandcost-effectivecomputationaltechniquesisagrowing necessitytoovercomethevastsearchspacerequiredtounderstand molecularmechanisms.Thisstudyisaconcertedefforttounder- standstructure-functionrelationshipsandexpressiondynamicsof CaHa-Trx-hprotein.Characterizingimportantproteinsimplicated indefenseresponsesofanagriculturallyimportantplantspeciesis theneedofthehour.
2. Materialsandmethods
2.1. Experimental
2.1.1. Plantandinsectgrowthconditions
Chickpeaseeds(C.arietinumL.;Pusa-362)wereprocuredfrom theIndianAgriculturalResearchInstitute,NewDelhi,India,were sowninpotscontainingautoclavedpottingsoilmixture(peatcom- postandvermiculite;1:1v/v).Plantsweregrownfor4weeksin a greenhouse with16/8h light/darkcycle at 22–25◦C, 50–60%
relativehumidity(RH),andwateredregularlyduringcultivation.
LarvaeofH.armigerawererearedinthelaboratoryat25◦Cand 65–70%relativehumidity(RH)ona14/10hlight/darkcycle.The larvaewerefedonanartificialdietasdescribedbyArmesetal.
(1992)[19].Thefreshlymoultedfifth-instarlarvaewerestarved over-nightbeforereleasingthemontheplants.
2.1.2. Planttreatments
Insectinfestationwasachieved bytherelease of fifth-instar H.armigeralarvaeon4-week-oldchickpeaplants(onelarvaper plant) and allowed to feed at 25◦C until 15–20% of the leaf areawasconsumed. Larvaewere thenremoved,and theentire shoot was harvested and stored at −80◦C after quick freez- ingin liquid nitrogen.To mimic insectinfestation, leaveswere wounded with a punch machine (hole diameter 44.5mm) (at 25◦C&65–70%RH.Plantsweresubsequentlyharvested.Fortreat- mentsinvolvingexogenoussignalingmolecules,equalvolumesof aqueoussolutionsofMeJA(100M;Aldrich,StLouis,MO,USA), SA(5mM;Sigma,StLouis,MO, USA),and ethephon(50M,2- chloroethanephosphonic acid, Sigma, St Louis, MO, USA) were sprayedontochickpeaplantsaccordingtopublishedprocedures [20]. Theplantswerethenharvestedand storedat−80◦Cafter quickfreezinginliquidnitrogen.
2.1.3. TotalRNAisolationandcDNAsynthesis
TotalRNA wasisolated usingRNeasyRNAisolation kit(Qia- gen)followingthemanufacturer’sprotocol.TheabsorbanceofRNA sampleswasdeterminedusingsmartspectrophotometer(Biorad).
RNAqualitywasmeasuredon1.2%formaldehydeagarosegeland bytheopticaldensity(OD)ratiosat260nm/280nm.Inorderto eliminateanygenomicDNAcontamination,totalRNAofeachsam- plesweretreatedwithDNaseI(Qiagen).RNAwasquantifiedusing
spectrophotometerandequal quantityofeach RNAsample was takenfor cDNA synthesis. Firststrand cDNAsweresynthesized usingRevertAidcDNAsynthesiskit(ThermoScientific)following manufacturer’sprotocol.
2.1.4. QuantitativeRT-PCR(qRT-PCR)andstatisticalanalysis ForrealtimePCR,thecDNAwasdiluted10timesandqRT-PCR wasperformedintriplicates inABIStepOneReal-time(Applied biosystems)usingSYBRGreenMasterMix(ThermoScientific)and gene-specificprimers.PrimersforqRT-PCRanalysisweredesigned usingthe primer3v.0.4.0 software[21,22] from uniqueregions.
Ampliconlengthswerekept50–150 basesforoptimalPCReffi- ciency. EachqRT-PCR reactionwasperformedin 10lreaction volumecontainingappropriatelydilutedcDNAastemplate,225nM ofeachforwardandreverseprimer,and2XPowerSYBRGreen PCRmastermix.Thermalcyclingconditionswere95◦Cfor2min followedby40cyclesof95◦Cfor15s,60◦Cfor1min.Therelative quantificationmethod(-CT)wasusedtoevaluatequantitative variationbetweenthereplicatesexamined.Theamplificationof actin2wasusedasanendogenouscontrolfornormalization.Sta- tisticalsignificanceofthedifferentialexpressionpatternsbetween treatmentswasdeterminedusingtheStudent’st-test.Forconsid- eringageneasHelicoverpa-orwounding-responsiveorsignaling compounds-responsive,thecriterionofminimum2-foldexpres- sionchange(atleastatonetime point)withP-value,0.05 was applied.
2.1.5. ExpressionpatternsofcaHa-Trx-hindifferenttissuetypes ForexpressionanalysisofCaHaTrx-hindifferenttissuesand organs during development, expression data available for the correspondingmRNA(TC04611)wereretrievedfromtheChick- pea Transcriptome Database (CTDB) (http://www.nipgr.res.in/
ctdb.html)andagraphcorrespondingtothatdatahasbeengener- ated.
2.2. Insilicoanalysis
2.2.1. Sequencealignmentsandphylogeneticanalysis
The full protein sequence was obtained by translating the mRNAsequencewithaccessionnumberXM004500914.1andis referredasCaHa-Trx-h (CicerarietinumplantcDNA)throughout themanuscript.TheanalysisofCaHa-Trx-hproteinwasperformed byProtParamtoolofEXPASYserver[23].Thehomologysearchwas carriedoutusingBLASTpprogramofNCBI[24]andsequencealign- mentwasperformedusingMUSCLEsoftware(v3.8.31)[25].Web basedtoolssuchasTargetPv1.1[26],MitoProt[27],andYLoc[28,29]
wereusedtopredictitsputativecelllocalization.Prodistprogram ofPHYLIPversion3.6a3[30]wasusedforstudyingphylogenetic relationships.Theunrootedtreewasmadeusingneighbor-joining (NJ)methodandplottedusingDrawTreeversion3.695ofPHYLIP.
2.2.2. Understandingfunctionalinteractivepartnersof thioredoxinhproteinfromA.thaliana
Afterascertaining the similarityof CaHa-Trx-h protein with Thioredoxin hproteinofA. thaliana,it iseasierto assumethat CaHa-Trx-hmustshowsimilarityinfunctionandprotein–protein interactions.Asbothseemtobepartofthesamemachinery,they mustelicitthesameresponsetostressintermsofproteinexpres- sionandco-regulation.Todelvefurtherwestudiedprotein–protein associationnetworksofA.thalianaTrx-hproteinusingtheSTRING database,version10.5[31].Apartfromalreadyavailableexperi- mentaldata,knowledgeofknownpathwaysandcomplexesfrom otherdatabases,itincludesdatafromco-expressionanalysis,lit- erature search, gene orthology based knowledge transfer, and detectionofsharedselectivesignalsacrossgenomes.
Table1
SequencessimilartoCaHa-Trx-halongwiththeirlocalizationandactivesite.
Proteinentrycode Organismname Proteinlength Putativelocalization Activesitesequence
CAA78462.1 Arabidopsisthaliana 114 Cytoplasm CGFS
AAN76509.1 Brassicarapasubsp.Oleifera 133 Cytoplasm CGFS
CAA55399.1 Chlamydomonasreinhardtii 113 Cytoplasm CGFS
AAP33009.1 Citrusxparadise 123 Cytoplasm CGFS
BAE16559.1 Codonopsislanceolata 124 Chloroplast CGFS
BAC21264.1 Cucurbitamaxima 120 Cytoplasm CGFS
ABB53600.1 Eucalyptusgrandis 117 Chloroplast CGFS
EKX51767.1 GuillardiathetaCCMP2712 187 Extracellular CGFS
AAU93947.1 Helicosporidiumsp.exSimuliumjonesi 112 Cytoplasm CGFS
AAD33596.1 Heveabrasiliensis 125 Chloroplast CGFS
ACS96439.1 Jatrophacurcas 118 Cytoplasm CGFS
AAO16555.1 Leymuschinensis 131 Chloroplast CGFS
AAZ32865.1 Medicagosativa 117 Cytoplasm CGFS
AAY42864.1 Nicotianaalata 152 Cytoplasm CGFS
AFP49340.1 Oleaeuropaea 123 Cytoplasm CGFS
AAB51522.1 OryzasativaIndicaGroup 122 Cytoplasm CGFS
EEC45257.1 PhaeodactylumtricornutumCCAP1055/1 165 Extracellular CGFS
CAC36986.1 Pisumsativum 120 Chloroplast CGFS
AAL90749.1 PopulustremulaxPopulustremuloides 143 Cytoplasm CGFS
AAL26915.1 Prunuspersica 136 Cytoplasm CGFS
EMS21648.1 RhodosporidiumtoruloidesNP11 241 Cytoplasm CGFS
AEM42995.1 Siraitiagrosvenorii 121 Cytoplasm CGFS
CAB96931.1 Triticumaestivum 125 Cytoplasm CGFS
ABX79345.1 Vitisvinifera 126 Chloroplast CGFS
2.2.3. Homologymodelingandvalidation
Thethree-dimensionalstructureofCaHa-Trx-hwassystemat- icallyelucidatedbyonlinehomology-basedstructureprediction serverModWebversionr189[32].Thestructuraldataisthenstored inModBase[33].It worksbysearchingforthefunctionallyand structurallyclosestproteintemplate.Anumberofmodelsarecal- culatedif morethan one template isavailable inthe database.
However,onlyonemodel waspredictedincase ofCaHa-Trx-h.
Thetemplate searchiscarried outusingPSI-BLASTorposition- specificiterated BLAST search and is also usedfor fold assign- ment.ThebeststructureisselectedbasedonModPipeQualityscore (MPQS)and sequenceidentitywhich isdiscussedin theresults section.Thebestscoringmodelwasautomaticallyselected.The presenceofconservedstructurewasstudiedusingNCBIConserved Domainanalysis[34] onmodeled CaHa-Trx-hprotein.Anaddi- tionalvalidationwasdonebythePSVS[35]serverthatincludes anumberofotheralgorithmsnamely,DSSP(secondarystructure prediction)[36,37],MolProbity[38],PDBvalidation,Verify3D[39], andPROCHECK3.5.4[40].Anadditionalenergy-basedvalidation wasalsorunonProsaserver[41].
2.2.4. Ashortmoleculardynamicsbasedsimulationofmodeled caHa-Trx-h
Thethree-dimensionalstructureofCaHa-Trx-hwassimulated forashortdurationtodelveintoitsdynamicproperties.Amber- Tools16 [42] were used for carrying out this simulation using amberff14SBBforcefields.Beforetheproductionrun,theprotein wasenergyminimizedtocleanupthestructureandstabilizefor MDsimulations.Theminimizationwascarriedoutfor1000steps;
100stepsofsteepestdescentwhichlaterisswitchedtoconjugate gradientmethod.ThepairwiseGeneralizedBornSolventmethod wasused.Nocutoff wasusedtoruna non-periodicsimulation.
A convergencecriterion (of energy gradient)value of0.01 was applied.Theenergyminimizedcoordinateswerethenputupfor production runusing Bornimplicit solvent modeland noperi- odicity.Theoutputwaswrittentofileevery500steps.Langevin thermostatwasusedtoregulatethetemperatureofthesystemat 300Kwithinitialandfinaltemperaturesto300K.Thesimulation ranfor25nswitha2fstimestep.TheSHAKEalgorithmwasusedto constrainbondsinvolvinghydrogen.Thisshortsimulationwillhelp ustounderstandlocalmotionsintheoveralltopologyandfindout
highlyfluctuatingregions.ThetrajectorywasvisualizedbyVMD anditssecondarystructurecalculatedusingits“Timeline”mod- ule.ThePrincipalComponentAnalysis(PCA)wasdoneusingthe cpptraj[43]moduleofAmberTools16.Theporcupineplots were createdusingVMDs[44]Nmwizutility.ThePCAplotwasmade usingMDtraj[45]toolsaccessedthroughijupyternotebook.
2.2.5. MechanismofactionthroughputativeArabidopsis thioredoxinHtargets
Toascertainthefunctionalaspectsofthenewlyreportedpro- tein,itsthree-dimensionalstructurewasdockedwithtwoknown targets of Thioredoxin h in Arabidopsis. These two targets are mannitoldehydrogenase(ManD)andPathogenesis-related5pro- tein(thaumatin).Duringpathogenattack,plantsexhibitexcessive oxidativestress,whichisdodgedbysomefungalpathogensdue tothe production ofmannitol, a well-known ROS quencher.In turn,plantsproducemannitoldehydrogenasetocatabolizemanni- tolproducedbyfungalpathogensandtherebymakingitsusceptible todamageduetoreactiveoxygenspecies(ROS)[46].Thelatterpro- tein,asthenamesuggests,isdirectlyimplicatedinplantdefense andbelongtothethaumatinfamily.Bothofthemhavebeenlisted asputativetargetsinMarchandetal.,2004[46]amongmanyothers butwechosethesegiventheirrelevanceinplantdefensesystems.
Also,theothersimilartargetsarenotsowell-knownandhence anappropriatehomologoustemplatewasn’tfoundforstructure modeling.
Since their experimentally determined three-dimensional structures were not available (forC. arietinum), we resorted to homology-basedmodeling using Modbase,an online serverfor Modeller.With71%and50%identitysharedwithcorresponding templatesfor ManDand thaumatin,respectively,thestructures wereobtainedand analyzed. Asa highly homologoustemplate forThaumatinwasn’tfound,wealsomodeleditsstructureusing I-TASSER,anabinitiomodeling server.Theproteinmodelsthus obtainedwerevalidatedusingPSVSasdescribedbefore.Thesepro- teinsweredockedwithCaHa-Trx-husingHaddockserver[47].The activesiteresidueswerechosentobecysteinesfromallthreepro- teins.ThioredoxinshaveaconservedsiteCys40-Cys43thattakes partinreducing targetproteins. Theaveragebonddistanceofa disulfidebondinaproteinis2.05Å,just0.5ÅmorethanaC C bond.TheSatomsofadisulfidebondinatargetproteinarereduced
234 A.Singhetal./InternationalJournalofBiologicalMacromolecules109(2018)231–243
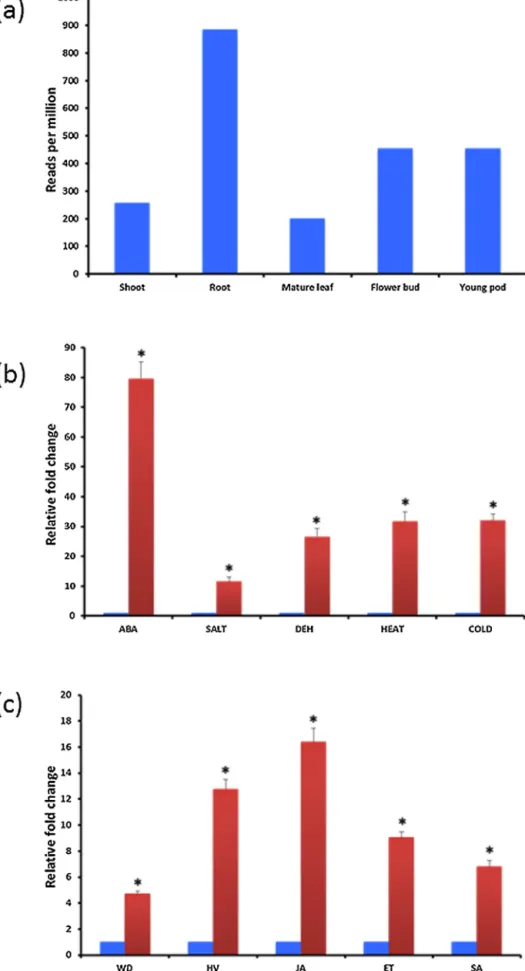
Fig.1.(a)ExpressionAnalysisofCaHaPR-4Aindifferenttissuetypes.(ExpressiondatahasbeenretrievedfromCTDB);(b)ExpressionanalysisofTrx-hduringdifferenttypes ofabioticstressesandABA,(c)ExpressionanalysisofCaHaPR-4AduringHelicoverpa-infestation(HV),Wounding(WD),SalicylicAcid(SA),MethylJasmonate(MeJa)and Ethephon(ET)treatments.
Fig.2.PhylogeneticrelationshipsofselectedThioredoxinsequencesfromdifferent plantspecies.
andthebondisbroken.Twocysteinesformingadisulfidebondis collectivelycalledacystineresidue.
3. Resultanddiscussion
3.1. InsilicoanalysisofcaHaTrx-h
ThepartialcloneofCaHa-Trx-hwasisolatedfromHelicoverpa- infestedChickpeaLibrary.Thefull-lengthmRNAsequenceandCDS wasutilizedforfurtheranalysis.ThisgenehasanORFof342bp andthededucedproteinsequenceof113aminoacids.CaHa-Trx-h hasthemolecularweightof12.39kDaandtheoreticalpIof5.67.
Thegrandaveragehydropathicity(GRAVY)valueis0.033andthe instabilityindexis13.05,whichsignifytheprotein’sstability.
CaHa-Trx-hshowedhighesthomologywith1XFLchainA(Solu- tionStructureOfThioredoxinH1)ofArabidopsisthaliana,1TI3chain A(SolutionStructureOfTheThioredoxinH1FromPoplar,ACppc ActiveSiteVariant)ofPopulustremula,1WMJchain A(Solution StructureOfThioredoxinTypeHFrom)ofOryzasativa,2VLTchain A (Crystal Structure Of Barley Thioredoxin H Isoform 2 In The OxidizedState)ofHordeumvulgare.Therespectiveproteinentry code,proteinlengthandputativelocalizationhavebeenshownin Table1.TheCaHa-Trx-hwaspredictedtohaveacytoplasmiclocal- ization.AccordingtotheclassificationofplantH-typesubfamily
‘FLKVDVDE’motifhasbeencharacterized.Another‘VVDFTASWCG- PCRFIAPIL’regionwasidentifiedasthemotifregioninCaHa-Trx-h.
3.2. ExpressionanalysisofcaHa-Trx-h
Expression patternof CaHa-Trx-hmRNAwas analyzedusing qPCR(Fig.1).Inthewholeseedling,moderatelevelsofCaHa-Trx- hmRNAexpressioncanbedetectedinnormalgrowthconditions, indicatingitsrequirementinthenormaldevelopmentalprocess (Fig.1a).Thesteady-statetranscriptofCaHa-Trx-hquicklyreached toahigherlevel(twelvefold)withinthreehoursofHelicoverpa- feeding.Duringmechanicalwounding,CaHa-Trx-htranscriptlevel increased(uptofourfold)butnotuptothatextentasthatofinsect feeding.Signalingcompounds(JA,SAandEt)playthecrucialrolein plantdefenseagainstpestandpathogen.Therefore,westudiedthe expressionlevelofCaHa-Trx-hmRNAaftertreatmentwiththese compounds.AccumulationofCaHa-Trx-htranscriptwasobserved atgreaterlevel(sixteenfold)duringJA-treatmentandatamod- eratelevelduringEt-treatment(eightfold)andSA-treatment(six fold)(Fig.1b).Thecriticalroleofthioredoxinsduringbioticstress hasbeenstated[10]anditwasshownthatTrxscatalyzesthecon-
Fig.3.(a)Anetworkofco-expressedandregulatedproteinsinrelationtoArabidop- sisthalianaTRX-Hprotein,(b)Adiagonalmatrixdescribingtheconfidencescoreof co-expressionoftwoproteins.
versionofcomplexNPR1tomonomer[48,49],whichisrequired forplantdefense.
Theinvolvement of thioredoxinsis welldocumentedduring differenttypesofabioticstresses[50,51,52].Consideringthat,we examinedthechangeinitsexpressionlevelduringABA-treatment, highsalt,dehydration,heat-stressand cold-stressandobserved thatmRNAlevelincreasesthirty-foldduringheat-stressandcold stressbuttoalesserextentduringdehydration(25fold)andsalt stress(10fold).TheCaHa-Trx-htranscriptaccumulationlevelwas foundtobeatthemaximumduringABA-treatment(Fig.1c).These results clearly indicate the potential role of CaHa-Trx-h during abioticstress.Therearemanyreportssuggestingtheroleofthiore- doxinsduringabioticstress.Over-expressionofAtTrx-h3confers heatresistancedue toitschaperonefunction[50,51].Similarly, OsTRXh1waswitnessedtobeinducedbysaltstressandcoldstress [52].
3.3. PhylogeneticanalysisofcaHa-Trx-hprotein
Toinvestigatephylogeneticrelationshipsof CaHa-Trx-hwith proteinsofotherplants,algaeandothereukaryotes,anunrooted phylogenetictreewasconstructed.Thetreewassub-dividedinto fivedifferentclades:Streptophyta,Bacillariophyta,Fungi,Chloro- phytaandpyrenomonadales(Fig.2).Thelargestcladeincluding CaHa-Trx-h of Cicer arietinum, Streptophyta comprises of Dicot and few Monocot plants like Arabidopsisthaliana, Brassicarapa subsp.Oleifera,Citrusxparadise,Codonopsislanceolata,Cucurbita maxima,Eucalyptusgrandis,Heveabrasiliensis,Jatrophacurcas,Ley- muschinensis,Medicagosativa,Nicotianaalata,Oleaeuropaea,Oryza sativaIndicaGroup,Pisumsativum,PopulustremulaxPopulustremu- loides,Prunuspersica,Siraitiagrosvenorii,Triticumaestivum,Vitis vinifera.SequencesfromRhodosporidiumtoruloidesNP11andGuil- lardiathetaCCMP2712showed42.94%and59.50%divergencewith theprotein,respectively.Thus,bothsequenceswereplaced dis- tantlyfromCaHa-Trx-hinthephylogenetictree(Fig.2).Theclose similarity ofCaHa-Trx-H withMedicago sativais notsurprising (bootstrapvale97outof100)asbothbelongtotheFabaceaefam- ilywhileitssimilaritywithpeach(Prunuspersica-bootstrapofonly 28),adeciduoustreeisquiteopposite.ThenextcladecontainsA.
thalianaandJ.curcas(bootstrap51),whichareofhighimportance asmodelplants.Thisclosesimilaritycanbeexploitedtomodel thioredoxinsystemsinanA.thalianasystemtobeappliedforcrop plantslikeC.arietinum.Citrus,EucalyptusandPisumsativumarethe restthreemembersoftheclosedclade.
236 A.Singhetal./InternationalJournalofBiologicalMacromolecules109(2018)231–243
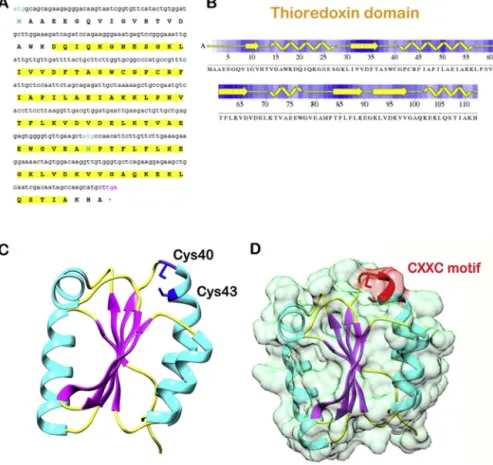
Fig.4.(a)ThetranslatedsequenceofpredictedCaHa-Trx-h(primarystructure),(b)secondarystructureofthesequence,(c)Thethree-dimensionalstructureofCaHa-Trx-h proteinwithhighlightedsecondarystructureelements,Cys40andCys43aremarkedinblue,(d)thesurfaceviewofproteinwithactivesiteCXXCmotifpresentatthesurface.
(Forinterpretationofthereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
3.4. Perspectivefromtheclosestrelative:A.thalianathioreodinh [PDB:1xfl]
Tounderstandtheprotein–proteininteractionnetworkofCaHa- Trx-h,agreatinsightcanbegainedbyobservingA.thalianaTrx1 network.Apreliminarysearchagainstthedatabaseshowsitsdirect interactionwith three proteins: NADPH-dependent thioredoxin reductases(NTRC, NTRA,and NTRB),glutamate synthases (GLT, GLU1,and GLU2),andperiredoxins(periredoxin-2bTPX1,1Cys periredoxinPER1,and2CysperiredoxinsAT3G11andAt5g06290).
AgraphicalnetworkisshowninFig.3a.Outofthese,thestruc- turesofglutamatesynthasesareunknownasdepictedbysmaller spheres.Homologousrelationshipscanbeobserved(asdepicted bylightblueconnections)amongst eachgroupofthenetwork;
allglutamatesynthases,NTRAwithNTRBandNTRC,andAT3G11 withAt5g06290(bothare2Cysproteins).Theblackcoloredcon- nectionsdepictco-expressionandallexceptTRX1withTPX1are showntobeco-expressed.Thisisastrongindicationofcoordi- natedstress-regulatorymechanisms.It wasalsosuggested that TRX1sharesgeneneighborhoodwithNTRsandperiredoxinsexcept TPX1depictedbygreenconnections.Theotherconnectionssignify thatthedatawasobtainedfrombothexperimentalandcurated databases.Geneco-expressioninA.thalianaisdescribedindetailin Fig.3b.Theintensityofredcoloredsquaresonthetriangularmatrix signifiestheconfidencethattwoproteinsarefunctionallyassoci- ated.TRX1doesnotshowco-expressionwithanyotherproteinbut otherproteinscanbeseenshowingstrongco-expression.Gluta- matesynthases1and2,2CysperiredoxinsAT3G11andAt5g06290, andNADPH-dependentthioredoxinreductasewithAT3G11show ahighconfidencescore.TheSTRINGdataisavailableinFig.1and Fig.2ofSupplementaryInformation1.
3.5. ModelingandanalysisofcaHa-Trx-hproteinstructure
Theprotein-familysearchforCaHa-Trx-hshowedthepresence ofthioredoxindomain(19–110aminoacids-Fig.4aandb)withan E-valueof5.07e-36.Atotalof107residueswerefoundtohavecon- servedfoldingamongwhichalmostallresidueswereconservedin astretch,thus,provingthatonlytheterminalregionsareflexible.
OnlyoneCaHa-Trx-hmodelwaspredictedwithA.thalianaThiore- doxinh1asthetemplate[PDB:1xfl][53]owingto73%identity.
TheEvalue(expectvalue<0.0001)ofzeroindicatesthesignifi- canceofalignmentandhomologousrelationshipasreportedby NCBIPSI-BLAST.TheGA341(Modelscore)indicatesthereliabil- ityofthemodelbasedonderivedstatisticalpotentials.Ascoreof 1.00(higherthan0.7)isconsideredthereliablei.e.probabilityof correctfoldassignmentismorethan95%.TheMPQSisacompos- itescoreofsequenceidentity,coverage,e-value,z-Dope(Discrete OptimizedProteinEnergy;threshold<0)andGA341scoreswhose valueof>1.1isconsideredreliable.Thescorefor CaHa-Trx-his 1.99915andthuscanbeconsideredareliablemodel.Thez-DOPE valueis−1.86indicatingareliablestructureprediction.Theoverall C␣-RMSD(Root-Mean-SquareDeviation)iscalculatedtobe1.215Å andthepredictednativeoverlap(3.5Å)is0.952Å.
Themultiplesequencealignmentshowedmaximumconserva- tionintheregionof1-␣1-2-␣2-3-␣3-3-4-␣3,whichisvery similartothioredoxinfamily.InCaHa-Trx-h,thethioredoxinmotif wasfoundinthe2-␣2region(Fig.4).Itstartedatthestartpoint ofthe2andcontinuedtillthemid-regionof␣2.Betasheetsform thecoreregionoftheprotein.Alphaheliceslieonthesurfaceofthe protein.Firstthreebetastrandsareparallel(1,2and3)and theothertwoareantiparallel(4and5).Thestructureendswith alphahelixandcontainsotherhelicesbetweenthesheets(Fig.4c andd).
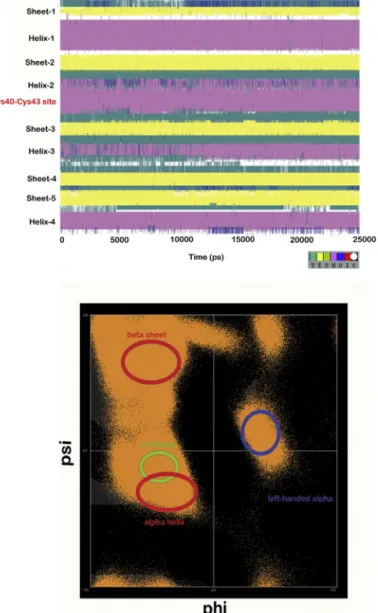
Fig.5.(a)TheevolutionofsecondarystructureofCaHa-Trx-HwithtimeduringMD simulations.TheactivesiteliesinHelix2,(b)Ramachandranplotofphi-psivalues ofeveryresidueaveragedforeachtimeframe.Theimportantsecondarystructural elementshavebeenhighlighted.
Inoneofthepreliminarystudiesonstructuralandfunctional characters of Thioredoxins by Eklund et al. [54], Asp26, Ala29, Trp31,Cys32,Gly33,Pro34,Cys35,Asp61,Pro76,andGly92were listed tobe conserved. The positions of theseresidues change slightlyinCaHaTrx-h.ThecorrespondingresiduesareAsp34,Ala37, Trp39,Cys40,Gly41,Pro42,Cys43,Asp68,Pro83,andGly99.These residuesformthesurfacearoundtheactivesiteandfacilitateinter- action.OtherobservedstructurallyimportantresiduewasPro40 whichcorrespondstoPro48whichintroducesa kinkin␣2and positionstheactivesiteproperly.Pro76isconsideredimportant formaintainingthestabilityofthestructure,whichcorrespondsto Pro83inCaHaTrx-h.
3.6. ValidationofcaHa-Trx-hmodel
ThestructurewasvalidatedusingRamachandraplotanalysis (Fig.3a-Supplementaryfile1).TheProcheckanalysisshowed94.9%
and5.1%ofresiduesinthefavoredandallowedregionsrespec- tivelywhilenoresiduewasdetectedasanoutlier.Similarresults wererecordedforRichardsonLab’sMolProbitywith99.1%and0.1%
ofresiduesinthefavoredandallowedregionsrespectively.The
GlobalQualityscoresfromdifferentmoduleswerecalculatedas follows:ProcheckG-factor(phi/psi)is0.20,Gfactor(alldihedral angles)is−0.01,andVerify3Dscoreis0.46.TheG-factorindicates thegoodnessofthemodel.Inotherwords,it’slog-oddsscorebased ontheobserveddistributionofstereochemicalparametersofpro- teinslikethedihedralanglesphi-psiandside-chainangles.These scoreshavebeenobtainedbyobservingtheseparametersin163 non-homologousstructuresobtainedbyX-raycrystallography.For aresidue,anegativeG-factorindicatesalow-probabilityconfor- mationandviceversa.Here,themeanG-scoreisgoodforphi-psi anglesbutnotforside-chainangleparameterswhichisalimitation incaseofcomputationalpredictionmethods.TheVerify3Dscore indicatesthecompatibilityofaproteinmodel(3D)withitsamino acidsequence(1D) onthebasisof thestructuralclassassigned toeachresidue (alpha,beta,loop,polar,non-polaretc).A posi- tivescoreof0.46(range−1∼badto+1∼good)indicatesanoverall good1D-3Dcompatibility.Noclosecontactswerefoundwithinthe rangeof2.2Å.TheRMSdeviationforbondanglesandbondlengths wasjust1.9◦and0.019Å,respectively.
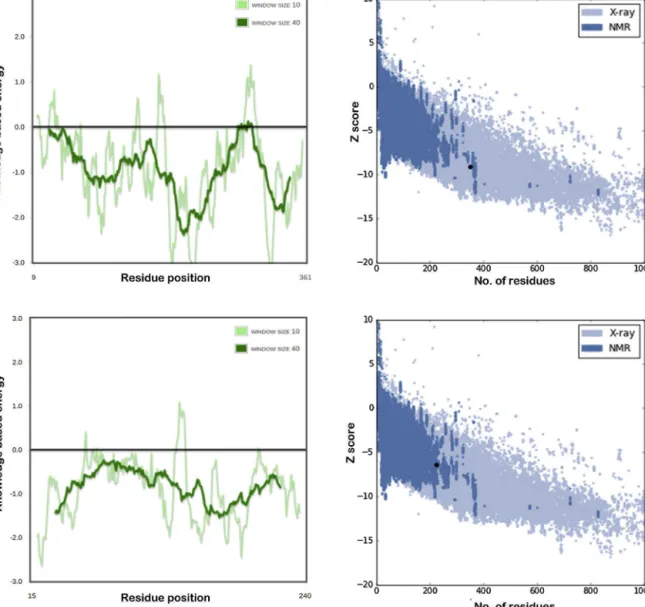
Anadditional runto validatethestructure by Prosayielded interestingresultsthatprovidesanoverallqualityscoreandcom- paresitwiththenativeproteinrange.IttakesonlyC-alphaatoms intoaccounttomakethiscomparison.TheZscoreofthiscalculation is−5.96andlieswithintherangeofexperimentallydetermined structures(Fig.3B-Supplementaryfile1).TheCaHa-Trx-hmodel doesnotdeviateintermsoftotalenergyofthestructure.Another energyplotinFig.3B(Supplementaryfile1)representslocalmodel qualitybasedonenergiescalculatedfor eachresidue. Theposi- tivevaluescorrespondtoerroneouspartsofthemodel.Toavoid largefluctuationsthatmayoccurduetosingleresidueplotting,a windowof10and40residuesisselectedforaccurateenergycal- culation.Thisexplainsaslightpositiveinclinationofenergyplot alonga10-residuewindowinFig.3B(Supplementaryfile1)which issignificantlyreducedinthe40-residuewindowplot.
3.7. Analyzingthestructurebasedonshortmoleculardynamics simulations
TheCaHa-Trx-hpredictedproteinwassimulatedforashorttime tostudymolecularfluctuationanddynamicstakingplaceona25ns timescale.Afteraninitialenergyminimizationwhichstabilizedthe structure,actualproductionoutputwasusedtocalculatethesec- ondarystructureevolutionovertime(Fig.5a)andaRamachandra plotwascalculatedbasedonaveragepositionsofdihedralangles phiandpsithroughoutthesimulation(Fig.5b).InFig.5a,thecol- orsdepict6differentsecondarystructuralelementsliketealfor turns(T),yellowforextended betasheets(E),greenforisolated bridge(B),magentaforalpha-helix(H),bluefor 3–10helix (G), redforpi-helix(I),andwhiteforrandomcoils(C).Thesecondary structureprofileofCaHa-Trx-hshowsastablebeta-sheetstructure throughoutthelengthofsimulationindicatingstabilitywhilethe helixregionsfluctuatetodifferentconformations.Acloseoverlap betweenalpha-helixand3–10helicescanbeseenforallhelical regions.Allhelicesareexposed tothesurfaceandfacestronger stericperturbationfromtheenvironment.Thesealsoformwhatis calledastheinteractivesitesliketheCys40-Cys43disulfidebond whichisfairlyexposed.Thedisulfidebondissurroundedbyflexible loopregionswhichimpartgreaterflexibilitytothearrangement witha potentialtarget.Italsoindicatestowardsapossibleshift instructureduetovaryingredoxstatesofCaHaTrx-h.InFig.5b, allimportantsecondarystructuralelementsaremarkedwithcir- cles.Aleft-handedhelixformationalsotakesplacealongwithfew non-secondarystructuralregionspopulatedbyProlineandGlycine residues.
RMSF(root-mean-square-atomicfluctuationforeachresidue averagedoverthetrajectory)andRMSDgraphsareshowninFig.6a
238 A.Singhetal./InternationalJournalofBiologicalMacromolecules109(2018)231–243
Fig.6. (a)Theroot-mean-square-fluctuationofeachresiduealongtheproteinchain,(b)radiusofgyrationalonganimaginaryaxisdepictingproteincompactness,(c)the Solventaccessiblesurfaceareaprofiledepictingtheareaincontactwiththesolventduringthesimulation,and(d)2-dimenionalmatrixofroot-mean-square-deviation ofbackbonecarbonatomsofallframes.Theredsquareindicatesthebiggestclusteroflow-RMSDstructuresformedtowardsendofsimulation.(Forinterpretationofthe referencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
andd,respectively.TheRMSDgraphisa2-dimensionalmatrixof deviationcalculatedovertime.ThebluecolorsshowlowRMSDand thus,highersimilarityandviceversaforregionsdepictedbyyel- low.Thematrixshowsthatthestructureexistsinfivestatesduring thesimulation.Thelargestclusterformsattheendofsimulation indicatingconvergence.
The Radius of Gyration (RoG) and Solvent Accessible Sur- faceArea (SASA)for thetrajectory areshown in Fig.6b and c, respectively.BothRoGandSASAplotssignifythattheproteinstruc- turewasmorecompactlyfoldedduringthesimulation,thereby,
decreasingthevalueofradiusofmoleculefromanarbitraryaxis thatpassesthroughthecentre-of-massofthestructure.Decreasing SASAvaluesalsosignifycompactpackingwithlessareaexposedto solvent.
Afteraninitialenergyminimizationwhichstabilizedthestruc- ture,actualproductionoutputwasstudiedusingtechniqueslike NormalModeAnalysis(NMA)[55]andPrincipalComponentAnal- ysis(PCA)[56].
Theanalysesshowthatthemostfluctuatingregionsofthepro- teinaretheN-andC-terminals.TheporcupineplotsinFig.7a,and
Fig.7.(aandb)Thetwomodesofmotiondepictedbycoloredporcupineneedles.Mostfluctuationcanbeseeninthesurfaceelementsandterminals.Theactive-sitedithiols arealsopresentatthesurfaceandarehighlightedwithcircles;(c)Principalcomponentbasedclusteringofvarioussnapshotsofthesimulationtrajectorybasedonrespective Cartesiancoordinates.Thestructuretraversesalargesearchspaceduringdynamictimeevolution.Thestructureseemstoexistsinfiveminimumenergyclusters,(d)A representativestructurefromeachclusterissuperimposedandtheactivesiteismarked.
bdepictthetwomodesofmotion.Thedirectionandmagnitudeof motionsisshownusingvaryingsizesofthepointers.Large-sized pointersdepictmajorfluctuationsandviceversa.Aconeextend- ingfromtheC-alphapositionofeachresidueshowsthedirection oftheatomalongtheeigenvector.Theterminalsarealmostnever conservedandshowhighdegreesoffluctuation(asdepictedbythe longersizeofpointers)whiletheproteincoreishighlyconserved and shows little orno fluctuation.The activesite Cys40-Cys43 (Helix2)wasalsoobservedwithalargemodeofmotion.Thedirec- tionofmovementofhelices2and3depictthatthesetworegions movewithrespecttoeachotherinanopen-closemannertofacil- itateapproachingtarget.Theresiduesthatforminternalcontact surfacebetweensecondarystructuralelementswereobservedby Eklundetal.[54],asPhe12,Val25,andPhe27whichcorresponds toVal33and Phe35whilethefirstPhenylalanineresidue isnot present.
AsimplePCAscatterplotasshowninFig.7cdepictsthespread ofdata(inthiscase the3Dcartesiancoordinatesofthesystem takenfromallframesofthetrajectory).Tounderstandthechanges undergonebythesystem,PCAisusedtoexplainthevarianceinthe coordinatespace.Theprincipalcomponents(PCsasinthex-and y-axes)depictthemodesofmotionandthefirstPCrepresentsthe dominantmotionandsoon.AplotofPC1versusPC2showstrajec- toryframesascoloreddata-points(afterdimensionalityreduction).
Thecolor-bardepictsthetimeinps.Notehowthepointsgraduate towardsacompletelydifferentconformationalsubspaceasafunc- tionoftime.Themostdominantmotiontakesplaceinthe10ns timestepwhilethesecondmostdominantmotiontakesoverafter atabout20ns.Italsodepictsthatthestructureexistsinabout5 energyminimaclustersanddifferentrepresentativestructurescan beattributedtoeachstage(Fig.7d).Itisunderstoodthattheactual
proteinmotionisdepictedasthesumofallindividualmodesof motion.Thesignificanceliesinthefactthatevenonananosec- ondtimescale;hugemolecularfluctuationscanoccurandgiverise totheplethoraofprotein–proteininteractionsandsignaling.This showsthatproteinsarenotrigidstructuresandapartfromastable coreregion,thesolvent-exposedregionsareconstantlychanging tointeractandbindwithtargetsormembranes.
Avalidationrunof theaveragestructureresultedinalower Z-scoreof−5.3fromProsaserver,whichsignifiesastructureof overallqualitywhencomparedwithexperimentallydetermined structures.
3.8. Modelingandvalidationofthree-dimensionalstructuresof ManDandthaumatin
Asdescribedearlier,duetotheabsenceofexperimentallydeter- minedstructures of thesetarget proteinsfor C.arietinum, they weremodeled andvalidated usingthesametools asfor CaHa- Trx-h. Thetemplatechosen forManDshowed71%identityand isanalcoholdehydrogenasefromPopulustremuloides.Thevari- ousscoresforthismodelareasfollows:E-value=0;GA341=1.00;
zDOPE=−1.13;RMSD=2.077;Nativecontactoverlap=0.94.Allthe calculations indicate towardsa reliable model and fold predic- tion. The other target Thaumatin wasmodeled using I-TASSER serverbecauseoflow-identitytemplatesearch.Itworksbytaking intoaccountmultipleprotein templatesthatshare highlysimi- lardomainsormotifswiththequery.Manytemplateswereused forthisprediction:ThaumatinfromThaumatococcusdanielli[2pe7, 2vu7, 3e3s, 2d8o], pathogenesis-relatedprotein from Nicotiana tabacum[1aun],anti-fungalZeatinfromZeaMays[1du5],Prunus aviumallergenprotein[2ahn-alsoselectedastheModwebtem-
240 A.Singhetal./InternationalJournalofBiologicalMacromolecules109(2018)231–243
Fig.8.Energyplotofthemodeledproteininwhichpositivevaluescorrespondtoerroneousparts.Thetwolinesdepicttwoseparatewindowlength.TheZ-scoreplot comparestheZscorewiththoseofexperimentallyknownproteins,(a)Mannitoldehydrogenase,(b)Thaumatin.
plate].Thecontinuousfragmentsobtainedbythreadingalignment are extracted and reassembled using replica-exchanged Monte Carlosimulations.SPICKERwasusedtoclusterthegenerateddecoy structureswithlow temperaturesandtop fiveclustercentroids areselectedforgeneratingfullatomicmodels.Outoffivehighly probablestructuresresultingfromthiscalculation,onewiththe highestCscoreandTMscoreisselected.TheC-scoreisameasure ofconfidenceinapredictedstructurebasedonthetemplatealign- ments.Itrangesfrom−5to2withahigherscoredepictinghigher confidenceinthemodel.ThepredictedthaumatinmodelhasaC- scorevalueof−1.38,whichisnotveryhighbutalsonotverylow.
AnothersuchvalueistheTMscore,whichimpartsabetterunder- standingofthedistanceofthepredictedstructurefromthenative.
Previousmethodslikeroot-mean-square-prediction(RMSD)can beambiguousduetolargeimpactoflocalerrorseveniftheglobal topologyiscorrect.ATM-score>0.5indicatesamodelofcorrect topologyandaTM-score<0.17meansarandomsimilarity.TheTM- scorevalueofthemodeledstructureis0.54+-0.15.Thethaumatin modelwasgeneratedfromaclusterwith3109decoysandaclus- terdensityof0.0564,whichisthehigherthanallotherclustersand signifiesampleoccurrenceofselectedstructureoverthecourseof simulationtrajectory.
Forvalidatingthesestructures,thePSVSresultsarementioned here.ForManDproteinmodel,theMolProbityRamachandrananal- ysispredicted96.9%ofitsresiduesfallinginthefavoredregionsand only0.6%inthedisallowedregions.TheGlobalQualityscoresfrom differentmoduleswerecalculatedasfollows:ProcheckG-factor (phi/psi)is0.00;Gfactor(alldihedralangles)is0.03,andVerify3D scoreis0.50,allofwhichindicatetowardsareliablemodel.Noclose contactswerefoundwithintherangeof2.2Å.TheRMSdeviation forbondanglesandbondlengthswasjust2.1◦and0.018Å,respec- tively.Forthaumatinmodel,MolProbityRamachandrananalysis predicted96.0%ofitsresiduesfallinginthefavoredregionsand only0.9%inthedisallowedregions.TheGlobalQualityscoresfrom differentmoduleswerecalculatedasfollows:ProcheckG-factor (phi/psi)is−0.36,Gfactor(alldihedralangles)is−0.27andthe Verify3Dscore is 0.46, all of which indicate towardsa reliable model.Noclosecontactswerefoundwithintherangeof2.2Å.The RMSdeviationforbondanglesandbondlengthswasjust2.3◦and 0.020Å,respectively.Outofallmodels,predictedthaumatinstruc- turehasthemostunreliablefoldprediction.TheN-andC-terminals showthehighestfluctuationandlessaccuracyduetotheabsence ofahighlyhomologousproteinofknownexperimentalstructure.
Thetwo structureswerealsovalidated usingthePROSA server
Fig.9.(a)CysteineresidueinteractionbetweenCaHa-Trx-handManDtargetprotein,(b)surfaceviewofCaHa-Trx-h-ManDcomplex,(c)residueinteractionbetween CaHa-Trx-handthaumatintargetprotein,(d)surfaceviewofCaHa-Trx-h-thaumatincomplex.
andresultedinenergeticallystableconformationscomparableto experimentallyknownstructures(Fig.8).
3.9. IdentificationofpotentialtargetsofcaHaTrx-hand understandingitsmechanismoffunction
Toelucidatethemechanismoffunctionofthemodeledpro- teinCaHa-Trx-h,itsinteractionpatternwithtwoputativeprotein targetsof AtTrxwasstudied.Theputativetargetsdescribed by Marchand et al., [48] implicated in defense mechanisms are (a) myrosinase,which is a glycoprotein and the glucosinolate- myrosinasesystemisa characteristicoffamilyBrassicaceae,(b) aposporyassociatedproteinC.Duetotheunavailabilityoffurther structuralknowledgeforArabidopsismyrosinaseandproteinC,we decidednottoselectthemforfurtherstudies.Theselectedpro- teintargetsaremannitoldehydrogenase(ManD)andthaumatinas identifiedforanA.thalianasystem.Thereductionmechanismof Trxsisadisulfidereducingsystemthroughawell-conserveddisul- fideactivesite(WCGPC).TovalidatethatCaHa-Trx-hisindeeda ThioredoxinhfromC.arietinum,weassumethatitshouldinter- actwith theselectedprotein targetsand reducethem through itsdisulfide(C C)bridge.Cys40-Cys43disulfidebridgeofThiore- doxinhhasbeenselected astheactive site basedon previous studies.Thedisulfidebridgeformedatthetargetproteinsurfaceis targetedtomonitorthisinteraction.TheHADDOCKscoreforCaHa- Trx-h-ManD interactionwas−74.4+/− 4.0. About182complex structureswerepredictedandgroupedinto11clusters.Thetop- rankedclusterhas41complexstructureswitha13.6+/−0.2RMSD fromtheoveralllowest-energystructure.Theenergieswerealso calculatedasfollows,VanderWaalsenergy:−28.1+/−2.9,electro- staticenergy:−162.3+/−22.2,desolvationenergy:−14.1+/−3.1, restraintsviolationenergy:2.4+/−0.54.Theburiedsurfacearea ofthecomplexis890.5+/−113.5.Lastly,theZscorevalueof−1.3 indicateshowmanystandarddeviationsthisclusterislocatedfrom theaverageintermsofthescore;themorenegativethebetter.The top-rankedstructurefromthis clusterwasselectedforanalysis.
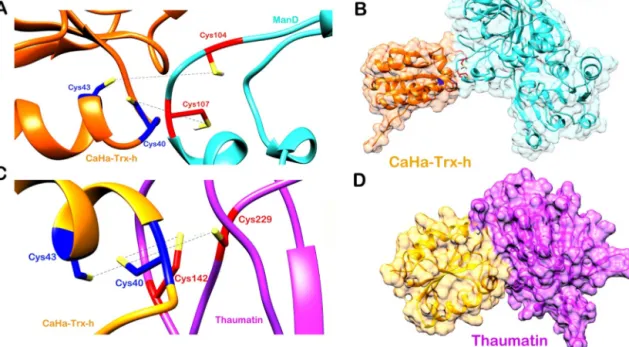
TheinteractionisdepictedinFig.9aandb.
TheHADDOCKscoreforCaHa-Trx-h-thaumatininteractionwas
−103.5+/−7.4.About175complexstructureswerepredictedand
groupedinto10clusters.Thetop-rankedclusterhas29complex structureswitha0.8+/−0.5RMSDfromtheoveralllowest-energy structure.Theenergies werealsocalculatedasfollows,Vander Waalsenergy:−49.2+/−3.8,electrostaticenergy:−211.3+/−47.0, desolvation energy: −14.6+/− 3.6, restraints violation energy:
26.3+/−17.76.Theburiedsurfaceareaofthecomplexis1390.9+/− 30.4.Lastly,theZscorevalueofthiscomplexstructureis−1.8.The top-rankedstructurefromthisclusterwasselectedforanalysis.The interactionconstitutesadirectlinkingofCys40-Cys43(CaHa-Trx- h)withCys142-Cys229(thaumatin)disulfidebridge(Fig.9cand d).
Comparing thetwo clusters,CaHa-Trx-h-thaumatin complex seemsmorestablethantheformerwithabetterHADDOCKscore andZscore.TheclusterRMSDfromtheaverageisalsoverylessin comparisontotheformer.Allenergyvaluesalsoindicateamore stableCaHa-Trx-h-thaumatincomplex.However,therestraintvio- lationenergyishigherthaninCaHa-Trx-h-ManDcomplex,which indicatesthatnotallrestraintswerefulfilled.Thismayalsoindicate alargerstructuralshiftduringinteractionanddisulfidebondbreak- ageleadingtobiggerviolations.Owingtoacorrelationobserved betweenbindingenergyandburiedsurfaceareabyChenetal.,2013 [57],itisnosurprisethatCaHa-Trx-h-thaumatincomplexismore stablethantheformer.Moreover,transcriptofPR-5showsinduc- tionduringHelicoverpainfestation[24],whichfurtherstrengthens ourpredictionthatCaHa-Trx-hinteractswithPR-5.
4. Conclusion
Thenecessityofpresent-dayworldtofindsustainableandratio- nalmethodstofightagainstvariousplantdiseaseshasledmany ofustodelvedeeperintoplantdefensemechanisms.Thioredox- insareanimportantclassofproteinsthatareubiquitouslyfound inthelivingworld.Amongstpartakinginothercrucialbiological processes,theirmostimportanttaskistoquenchtheROSstress.
Variousproteinscanexisteitherinoxidizedorreducedformdefin- ingdifferentstatesofactivity.Theoxidizedproteinsarereduced byThioredoxins.Trx-h,fromCicerarietinum,becamethesubjectof thisstudywhenitwasfoundtobeexpressedduringHelicoverpa infestation. Inthisstudy,CaHaTrx-h wasstudiedforitsexpres-