ContentslistsavailableatSciVerseScienceDirect
Journal of Plant Physiology
j o ur na l hom e p a g e :w w w . e l s e v i e r . d e / j p l p h
The taxon-specific paralogs of grapevine PRLIP genes are highly induced upon powdery mildew infection
Balint Szalontai
a, Szilvia Stranczinger
a, Gergo Palfalvi
a, Brigitte Mauch-Mani
b, Gabor Jakab
a,∗aInstituteofBiology,FacultyofSciences,UniversityofPecs,Hungary
bInstituteofBotany,UniversityofNeuchâtel,NeuchâtelCH-2009,Switzerland
a r t i c l e i n f o
Articlehistory:
Received5March2012
Receivedinrevisedform29June2012 Accepted2July2012
Keywords:
Arabidopsis Benzothiadiazole Ethylene Grapevine Powderymildew
s u m m a r y
PRLIP(pathogenesis-relatedlipase) isagenefamilyencodingclass 3 lipase-likeproteinsoriginally describedandfirstcharacterizedinArabidopsisthaliana.NineparaloggenesofArabidopsiscanbesep- aratedintotwogroupsbasedonexpressioncharacteristicsandpathogenresponses.GenesofGroup1 areclusteredonchromosome5andshoweitherhighinducibilitytodifferentstresshormonesandin responsetopathogenattackorareundetectableatthetranscriptlevel.Group2containstheremain- inggenes,spreadoverthegenomeandareexpressedconstitutivelyinallthetissuestested.Theaim ofthepresentstudywastodeterminethedistributionofthesetwogroupsamongplants,andtover- ifytheirdifferentialexpression.Orthologsofconstitutivelyactivemembers(Group2)werefoundin allangiosperms,withavailablegenomesequences.Theyarereferredtoas“corePRLIPs”.Incontrast, thegeneclustercontainingthepathogen-induciblePRLIPs(Group1)wasuniqueforArabidopsis.Among otherangiosperms,grapevinealsopossessessuchauniquegenome-specificgroupofPRLIPgenes.To investigatewhetherthesegenesarealsocounterpartsinpathogenresponses,theirexpressionpattern wastestedunderstressconditions.TwoofthespecificVitisPRLIPswerehighlyinducedinresponseto bothpowderymildewinfectionandbenzothiadiazole(BTH)treatment.CoreVitisPRLIPs,however,were notresponsivetoeitherpathogenattackorthechemicalinducer.Ourdataprovideinsightsintothe distributionofapathogenesis-relatedgenefamilyindifferentplantlineages,andmightrevealcommon characteristicswithotherinducibledefense-relatedgenefamilies.
© 2012 Elsevier GmbH. All rights reserved.
Introduction
PRproteinsareimportanteffectormoleculesofplantdisease responses.Theyarecommonlyfoundintheplantkingdomandare currentlyclassifiedinto17PRfamiliesbasedonbiologicalandbio- chemicalproperties(VanLoonetal.,2006).However,noPRfamily consistingof lipasesor lipase-like proteinshasyetbeen estab- lished,althoughrecentstudieshighlighttheimportanceoflipases andlipidicsignalsindifferentplantstressresponses(Shah,2005).
Lipases,alongwithesterases,belongtothealpha/beta-hydrolase foldfamilyofenzymes.Theypreferentiallyhydrolyzelong-chain acylglycerolsintoglycerolsandfree fattyacids,whilethelatter primarilyhydrolyzeshortchainacylglycerols(Ollisetal.,1992).
Thestresshormonejasmonicacid(JA),alipidderivativeitself, hasbeenproposedtoplayaroleinlipidicsignalinginJA-mediated
Abbreviations:BTH,benzothiadiazole;CDS,codingsequence;PR,pathogenesis- related;RQ,relativequantitation;SAR,systemicacquiredresistance.
∗Corresponding authorat: University of Pecs, Ifjusag utja 6, PecsH-7624, Hungary.Tel.:+3672503600x24209;fax:+3672503634.
E-mailaddress:jakab@gamma.ttk.pte.hu(G.Jakab).
woundresponses(FarmerandRyan,1992).Salicylicacid(SA)medi- atedSARisalsostronglyaffectedbylipidicsignalingthroughSABP2, aproteinhavingSA-stimulatedlipaseactivity,andthusactingas a receptorfor SAintobacco(KumarandKlessig,2003).InAra- bidopsis,asimilarfunctionhasbeenattributedtothehomologous AtMESproteinswithesteraseactivity(Vlotetal.,2008).Onegroup ofstress-relatedlipasesofspecialinterestistheGDSL(alsoknown asSGNH)lipases.GLIP1,aGDSL-typelipase,wasobservedtobe highly induced inresponse to ethephontreatment and toplay animportantroleininducibleresistanceagainstAlternariabras- sicicola.Itmightbeimplicatedineitherthesignalingprocessor exertantimicrobialactivity(Ohetal.,2005).Overexpressinglines ofGLIP1exhibitedincreaseddefenseagainstAlternariaandwere assumedtoformaGLIP1-ethylene(ET)signalingpathwayfunction- ingindependentlyfromSAR(Kwonetal.,2009).Anothermember oftheArabidopsisGLIPgenefamilyGLIP2hasalsobeenreported toaffectdefenseresponsesthroughrepressionoftheauxinsig- nalingpathway.However,thetranscriptlevelsofGLIP2werealso enhancedbySA,methyljasmonate(MeJA)andethephon(Leeetal., 2009),suggestinginvolvementinmultiple,evenantagonisticsig- nalingpathways.AGDSLlipasefromhotpepper (CaGLIP1)with anorgan-andtissue-specificexpressionpatterndisplayedreduced 0176-1617/$–seefrontmatter© 2012 Elsevier GmbH. All rights reserved.
http://dx.doi.org/10.1016/j.jplph.2012.07.010
etal.,2008).OverexpressionofCaGLIP1causedenhanceddisease susceptibilityintransgenicArabidopsisplants.AnotherGDSLlipase frompepperwashighlyinduciblebyMeJAaswellaswounding, andisalsolikelytomodulateCaPR4expression(Kimetal.,2008).
Br-sil1,aputativelipasefromChinesecabbageleavesandstems (LeeandCho,2003),ishighlyinduciblebytheSAanalogbenzoth- iadiazole(BTH)andbyPseudomonassyringaeinfection,butneither byJAnorbyET.TheGDSLlipaseGER1fromricecanbeinducedby JAandredandfarredlight(Riemannetal.,2007).
Class3lipasesarealsoinvolvedinplantstressresponses.The familyofalpha/beta-hydrolasescontainingclass3lipasedomains comprisesthreeproteinswithelementalrolesinstressresponses ofArabidopsis.TheseincludePAD4(Zhouetal.,1998),EDS1(Falk etal.,1999)andSAG101.Theyformasignalingsystempresumably functioningasthemainbarrieragainstpathogens(Wiermeretal., 2005).Despitetheirbiologicalcharacterization,notmuchisknown abouttheinvivolipolyticactivityandbiologicalsubstratesofthe class3lipases;itisactuallyuncertainwhethertheyexhibitsuch activity(Wiermeretal.,2005).ThePRLIPsarealsoagroupofclass 3lipasesoriginallycharacterizedbyJakabetal.(2003).Ninemem- bersofthegenefamilywereidentifiedintheArabidopsisgenome, andthreeof them–PRLIP3,PRLIP8and PRLIP9–werereported tohaveorthologsinthericegenome.ExpressionlevelsofPRLIP1, PRLIP2andPRLIP6werehighlyinducedbydifferentbioticstress stimuliwhile,PRLIP3,PRLIP9andPRLIP8genesshowedonlymod- erateorlowbioticstressresponses(SzalontaiandJakab,2010).
Theaimofourpresentstudywastohighlightfunctionaldifferenti- ationbetweenPRLIPs,asindicatedinArabidopsis.Theinformation presentedhereis important inunderstandingthephysiological functionofthePRLIPs,whichmaybeanewfamilyofPRgenes.
Materialsandmethods
Genomicdatabasesearchandphylogenicanalyses
Databasesof25sequencedandannotatedplantgenomesavail- ableinthePhytozomev7.0Database(http://www.phytozome.net) werescreenedtoidentifypotentialPRLIPhomologsequences.To improvethealignment,insomecasescomputationallypredicted exon–intronboundarieswererevisedtoresultinbetterhomology attheaminoacidlevel.SequenceswerealignedwiththeJalView programusingtheMUSCLEmethod.Phylogeneticanalysiswascar- riedoutonly onthesatisfactoryalignedregion,usingPAUP*4.0 (Swofford,2003)withtheDistanceoption.Aphylogenetictreerep- resentingsimilaritiesofproteinsequenceswasconstructedusing theneighbor-joiningmethod.Bootstrapsupportsof1000replica- tionsareindicated,andthecut-offvalueforcondensedtreewas
<60.
Plantmaterialandhormonetreatments
VernalisedcuttingsofVitisvinifera(L.)CabernetSauvignonand PinotNoircultivarswereobtainedfromtheInstituteofViticulture andOenology,UniversityofPecs.Vine-stocksweretreatedwith 0.8% alpha-naphtalene acetic acid (INCIT-8; Bioplant, Hungary) androotedinperlite.Plantsweregrownat28◦Cunder16hlight period,70Em−2s−1 illuminationand60%relativehumidityfor 6weeksaftertheemergenceofbuds.Toinduceresistancemech- anisms,afreshsolutionofbenzo(1,2,3)thiadiazole-7-carbothioic acidS-methylester(Syngenta,Switzerland)atafinalconcentra- tionof 600M– dissolvedin tapwater – was appliedas soil drench.ETgastreatmentwasperformedinanairtightcontainer withaconcentrationof100LL−1.Grapevineleavesinfectedwith
pooled(10leavespersample).SamplesincludedPN0:uninfected leaves;PN2:5–10%;PN5:30–40%;andPN8:>75%correspondingto thesurfaceofvisiblyinfectedleafsurfaceinpercentage.Thenum- berofbiologicalreplicateswasn=3forchemicaltreatments,and n=2(percultivar)forpowderymildewdiseaseresponses.
RNAisolation,reversetranscriptionandquantitativePCR
GenomicDNA was purified fromleaveswiththe useofthe DNeasy Plant Mini Kit (Qiagen, Germany). Total RNA was iso- latedfromflashfrozenplanttissuesaccordingtotheprocedure ofHamiduzzamanetal.(2005).AfterDNaseI(Fermentas,Lithua- nia)treatment,thequalityoftotalRNAsampleswascheckedby separatingona1.2%agarosegelcontaining5%formaldehyde.RNA quantitiesweremeasuredspectrophotometricallyinaNanoDrop ND-1000spectrophotometer(ThermoScientific,USA).cDNAwas synthesizedfrom200ngoftotalRNAusingtheFirstStrandcDNA Synthesis Kit(Fermentas) witholigodT(18) primer followingthe manufacturer’sprotocol.QuantitativeRT-PCRswereruninareac- tionvolumeof20lconsistingof10lMaximaTMSYBRGreen/ROX qPCRMasterMix(Fermentas),1.0mMeachoftwoprimers,8.5l waterand2lofcDNAsampleonaStepOneTMReal-TimePCR System(AppliedBiosystems,USA),withthefollowingprogram:40 cyclesof95◦Cfor30s;60◦Cfor30s;and72◦Cfor60swithanini- tialdenaturationat95◦Cfor10min.Geneexpressionlevelswere calculatedbynormalizationrelativetoelongationfactor1-alpha mRNAlevel(GenBankaccession:XM002282094),asitdisplayed themoststabletranscriptioninthesampleswhencomparedto abeta-actinandtwotubulinetranscriptlevels(datanotshown).
PrimersequencesarelistedinSupplementaryTable1.
Statisticalanalysis
Forstudiesontheeffectsofchemicaltreatmentsandpowdery mildewinfection,eachleafsamplewaspooledfromshootsof5 differentcuttingsper time point.Whenstudyingorganspecific expression,eachsamplewaspooledfrom5 differentindividual plants.Datapresentedasbargraphsshowonerepresentativeof 3biologicalreplicatesofchemicaltreatmentsandorgan-specific expressionanalysis,andonerepresentativeof2biologicalrepli- cates(percultivar)ofpowderymildewdiseaseresponses.Samples weremeasuredintriplicateandrelativequantificationwasper- formedwiththeCT methodusingStep OneTM 2.0Software (AppliedBiosystems).Theefficiencyvaluesof theprimerswere determinedbyestablishing5-folddilutioncurvesandcalculated asdescribedbyPfaffl(2001).Accordingtotheoutputformatofthe software,results(RQvalues)aregivenasmeanswith95%confi- denceintervals(CIs).InFigs.3–6,barsrepresentmeansanderror barscorrespondtoCIs.Inthisgraphicalrepresentation,twomeans withnon-overlappingCIswereconsidereddifferentatthep<0.05 levelofsignificance.
CloningofPCRfragments
PCR productsfor cloning were generated withDreamTaqTM DNAPolymerase (Fermentas)ina 23lreactionvolume(2.3l DreamTaqTM Buffer, 0.5mM dNTP mix, 1.0M of forward and reverse primers, 4l of gDNA or cDNA template 1.25U DreamTaqTM DNAPolymerase and12.5lwater).PCR products were recovered from 1.5% agarose gels with the GeneJET Gel ExtractionKit (Fermentas) following thesupplier’s instructions.
AmpliconswerebluntedandclonedtothepJET1.2vectorusingthe GeneJETPCRCloningKit(Fermentas).EscherichiacoliDH5alpha competentcellsweretransformedasdescribedbySambrooketal.
Fig.1. PRLIPgeneclustersofthreeangiospermspecies.GreyboxesstandforPRLIPhomologs,blackboxesforothergenes(“c”:encodingGTPbindingproteinhomolog;“d1”
and“d2”:pseudogeneswithintheVitisPRLIPcluster)whiteboxesfornon-codingRNAgenes(“a”:At5g24205and“b”:At5g24206).Orientationsareindicated.Accession numbersarelistedinSupplementaryTable2.
(1989).TherecombinantcloneswereanalyzedbycolonyPCRwith pJET1.2forwardand reverseprimersaccordingtothemanufac- turer’sprotocol.DNAsamplesweresequencedusingtheBigDye TerminatorCycleSequencingKit(AppliedBiosystems).
Results
PlantPRLIPscanbedividedtocoreandgenome-specifichomology groups
Since the first characterization of the PRLIP gene family in the model plant Arabidopsis thaliana, the genome of Arabidop- sis lyratahas also been sequenced,so we compared the PRLIP gene cluster of thesetwo closely related species.We usedthe DOE-JointGenomeInstitutedatabase(http://www.jgi.doe.gov/)to comparePRLIPgenesofthetwoBrassicaceaespecies.Constitutively expressedmembers(asdemonstratedearlierinA.thaliana)PRLIP3, PRLIP8andPRLIP9arepresentinbothgenomes,locateddispers- edlyindifferentregions(datanotshown),whiletheremainderof thePRLIPgenesencodingthepathogeninduciblemembersinA.
thalianaoccurredinonegeneclusterinbothspecies.Thegenes areina collinearpositionalong thehomologchromosomeseg- ment,exceptforPRLIP2,whichismissingfromtheA.lyratagenome (Fig.1).Thereareadditional significantdisparitiesbetweenthe twospecies.InA.lyrata,thereisashortCDSbetweenPRLIP1and PRLIP4encodingaputativeGTP-bindingprotein,whichcouldnot belocatedinA.thaliana.Anotherexampleofinterspecificdiffer- encesisthenumberofPRLIP4copies.OrthologsofthePRLIP4areina collinearpositioninbothregionscompared,buttherearetwoaddi- tionalparalogsofthisgenethatarepresentonlyinA.lyrata.These
showsimilaritytoPRLIP4,butthe5regionismissinginbothgenes, sotheyarelikelytheresultofpartialduplicationsratherthanintact genes.Amissing5codingregionwasalsofoundinAlPRLIP1and AlPRLIP9(whencomparedtoA.thalianaorthologs).Finally,there aretwonon-codingRNAgenesintheA.thalianagenomebetween thePRLIP2andPRLIP1genes,whichareyetnotindicatedintheA.
lyratahomologsegmentbutcanbeidentifiedbyahomologysearch andarethuslikelytobepresentinbothgenomeclusters.
Despite the close phylogenetic relatedness, 5 million years of independent evolution (Lysak et al., 2006) were sufficient to develop remarkable disparity between PRLIP gene assort- ments of two Arabidopsis species. Therefore, we screened the proteome databases of the 25 plants available at Phytozome (http://www.phytozome.net)toidentifyhomologsofAtPRLIPpro- teinsandtoestimatesequence-levelsimilarityamongthem.After criticalrevisionoftheBLASTmatches,135sequencesfrom23plant specieswereconsideredtobeindisputablePRLIPhomologs.Our aimwastoidentifypotentialfunctionalgroupsofPRLIPproteins (whichmightactuallycontributetophysiologicalprocesses)and not toreconstructtheentire molecularevolution of themulti- gene family.Forthis reason,genomeloci withnointactcoding regionassigned andtruncatedproteinfragmentswereexcluded fromthefurtheranalysis.ThemossPhyscomitrellapossessesthree PRLIP sequencesthat were used as theoutgroup in theanaly- sis.TheingrouponthephylogenictreeconsistedofTracheophyte sequences (Fig. 2; for bootstrap supports, detailed tree topol- ogyandaccessionnumbersseeSupplementaryFig.1).Accessions of Selaginella appeared as a sister in the basal position, while angiospermsequencesformtwomainclades,referredtoasCladeI andCladeII.AvarietyofEudicotsequencesappearintheterminal branchesinbothoftheseclades:CladeIcontainingthehomologs
Fig.2.PhylogeneticrelationshipsofthePRLIPproteinsequences.Boldlinesindicategenome-specificparaloggroups.NotethatspecificgroupsinBEPgrassesandPACCMAD grassesshouldbeinterpretedseparatelywithineachspecies.Seetextforinterpretationofbranchesmarkedwith“a”and“b”.
ofAtPRLIP8,andCladeIIcontainingthehomologsofAtPRLIP3and AtPRLIP9.Inaddition,fourparalogsforPRLIP8andtwoforPRLIP3/9 arepresentingrasses,inferringageneduplicationeventinoneof thecommonancestors.Allofthesequencedplantshaveofatleast onecopyofPRLIP8andPRLIP3/9,sowetermthemcorePRLIPs.
Interestingly,therestofthesubcladesinCladeIIconsistmostly ofaccessionsfromasinglegenomeeach.UnlikethecorePRLIPs, thesegenome-specificparaloggroups containproteinsencoded inonegenecluster,withtheexceptionofsubcladesofMimulus andGlycine.Altogether,suchgenome-specificsubcladesofPRLIPs couldbeestablished in 9 of the23 analyzed species, and only one(including twoMedicagoaccessions)islocatedwithinClade I.Nevertheless,there areunique PRLIPsequences,dissimilar to theircorePRLIPs, in Brachypodium,Manihot,and Citrus (one in eachspecies,formingpolyphyletic cladeswithOryzaaccessions andeachotherrespectively–markedas“a”onFig.2).Thepre- viouslydescribedandcharacterizedA.thalianaPRLIPsclusteredon chromosome5(includingPRLIP1,PRLIP2,PRLIP4,PRLIP5,PRLIP6 andPRLIP7;Jakabetal.,2003)allbelongtooneofthesespecific paraloggroups, and thus are singular in theArabidopsisgenus.
Remarkably,PRLIP8sequencesofMimulus,togetherwithaunique sequencefromGlycine(whichispredictedtobeapartialduplica- tion),divergedfromtheothercoreeudicotPRLIP8sequencesand wereexcludedfromtheterminalnode(markedas“b”onFig.2).
Mostofthesequencedataarepredictedproteinsassignedtohypo- theticalcodingregionsandmightincludeseveralpseudogenes.
Coreandgenome-specificPRLIPgeneshavedifferentexpression patternsingrapevine
Arabidopsis PRLIPs show a characteristic expression pattern dependingonwhethertheybelongtothecoreorgenome-specific groups (Jakab et al., 2003; Szalontai and Jakab, 2010). In our additionalexperiments,weanalyzedthePRLIPgene assortment ofgrapevine(VvPRLIPs)todemonstratethiscorrelationbetween sequence-levelsimilaritiesandfunctionaldifferentiationinadis- tantlyrelatedplantspecies.AsgenepredictionsoftheGenoscope grapevinegenomedatabase(http://www.genoscope.cns.fr/spip/) seemedtobepoorfortheVvPRLIPs,weusedannotationsoftheNCBI MapViewer(http://www.ncbi.nlm.nih.gov/projects/mapview/)for further analyses. A single copy for both core PRLIPs (desig- natedasVvPRLIP8and VvPRLIP3/9) canbeidentifiedwithinthe Vitis genome.In addition, a gene cluster containingduplicated genesof the Vitis-specific PRLIPsis located onchromosome 16 (Fig.1).Altogether,sevenhomologgene lociin this regiondis- playedhomologytoAtPRLIPgenes(termedasVvPRLIPE,VvPRLIPA, VvPRLIPF,VvPRLIPC,VvPRLIPG,VvPRLIPDandVvPRLIPBtodistinguish themfromArabidopsis-specificPRLIPgenes).ThelocusVvPRLIPGis apparentlyagenefragment.Manualrevisionoftheexon–intron junctionswasneededtoimprovehomologytootherVvPRLIPsfor VvPRLPCandVvPRLIPF(forproteinalignment,seeSupplementary Fig.2;accessionnumbersarelistedinSupplementaryTable2;for manuallycurated,sequencesseeSupplementaryTable3).
0 50 100 150 200 250 300
R S L F
RQ
VvPRLIPA
A
0 2 4 6 8 10 12
R S L F
RQ
VvPRLIPC
B
0 2 4 6 8 10 12
R S L F
RQ
VvPRLIP3/9
C
0 2 4 6 8 10 12
R S L F
RQ
VvPRLIP8
D
Specific PRLIPs Core PRLIP s
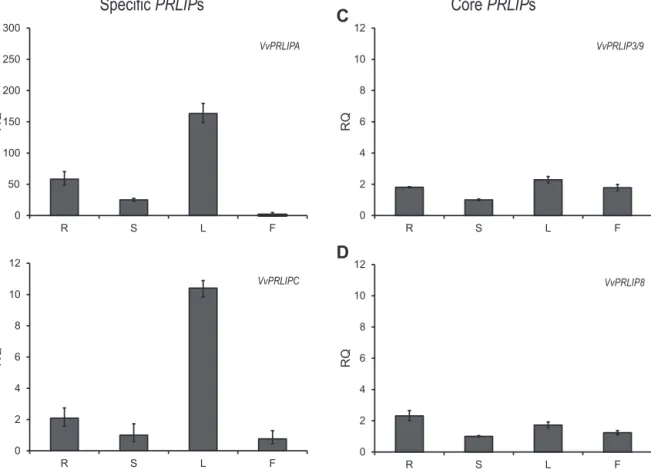
Fig.3. RelativebasalexpressionofgrapevinePRLIPgenesamongdifferentplantorgansinthePinotNoircultivar.R,root;S,stem;L,leaf;F,flower.Errorbarscorrespondto technicalreplicates.
First,weidentifiedgenesthatactuallyappearatthetranscript level.Byexploitingthehighlevelof sequencesimilarityamong theclusteredVvPRLIPs,wedesignedauniversalprimerpair(VvU- NIV) able to amplify all of the predicted specific VvPRLIP loci.
This capacity of the primers first was tested on Vitis genomic DNA. Sequence analysesconfirmed that amplicons of allseven homologlocifromthegrapevinePRLIPclustercouldbeobtained.
Next,VvPRLIPfragmentsfromcDNAsamplesofbothinfectedand uninfected grapevineleavesand alltheotherexaminedtissues wereamplified.Thesefragmentswereclonedand sequencedas describedin ‘Materialsand methods’section.Atleast30 clones representingeachsamplewereanalyzedandourresultsindicated thatonlytwoparalogs(VvPRLIPAandVvPRLIPC)weredetectableat themRNAlevelinthegrapevineorganstested(datanotshown).
Notably,itispossiblethatothermembersofthegeneclusterare alsotranscribedincertaincelltypesortissues,orunderspecial conditions.Furthermore,competitionbetweenthedifferenttem- platesfortheuniversalprimermightalsooccur,favoringVvPRLIPA andVvPRLIPCtranscripts.
Todeterminepotentialdefense-relatedcharacteristicsofthe genefamily, wealsostudiedtheexpressionprofilesoftheVitis PRLIPorthologs.Inuntreatedgrapevineplants(cultivarPinotNoir), corePRLIPsdisplayedonlysmalldifferencesinexpressionamong thecomparedtissues(Fig.3CandD),withonly2.5-foldatthemax- imum.Conversely,specificPRLIPsVvPRLIPAandVvPRLIPCshowed veryhighlevelsofexpressioninleaves(Fig.3AandB)comparedto otherorgans.
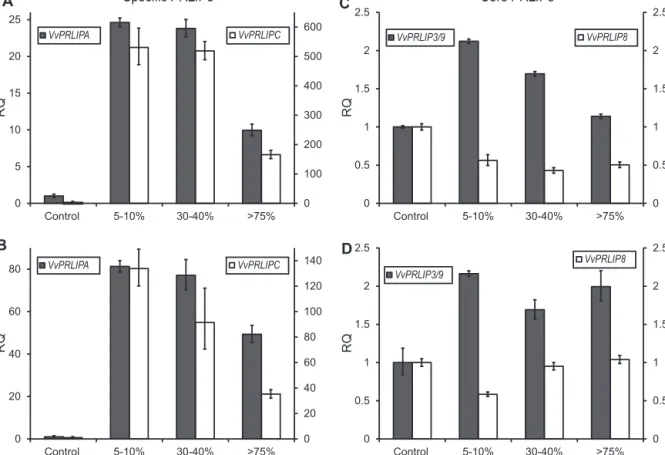
Wealsostudiedexpressionchangesofthesegenesinresponse todifferentstressregimens.ComparingVitisleavesunderdiffer- entratesofpowderymildewinfectionrevealedsignificantchanges in the mRNAlevels of VvPRLIPA and VvPRLIPC(Fig.4A and B).
Low levels of infection caused 25-fold (Pinot Noir) or 80-fold
(Cabernet Sauvignon) upregulation in VvPRLIPA transcript lev- els,while VvPRLIPCdisplayed 500-foldand more than125-fold increases in the same infected leaves of the two cultivars, respectively. Incontrast, asevere infectionmarkedly decreased expressionlevelsofgrapevine-specificPRLIPgenes.Forthecore VvPRLIPs,only minordifferences weredetected(Fig.4Cand D):
VvPRLIP8showed0.5-folddownregulationinmoderatelyinfected leavesofPinotNoir,whereas VvPRLIP3/9displayedonlya weak induction,withamaximumof2-foldincreaseinbothcultivars.
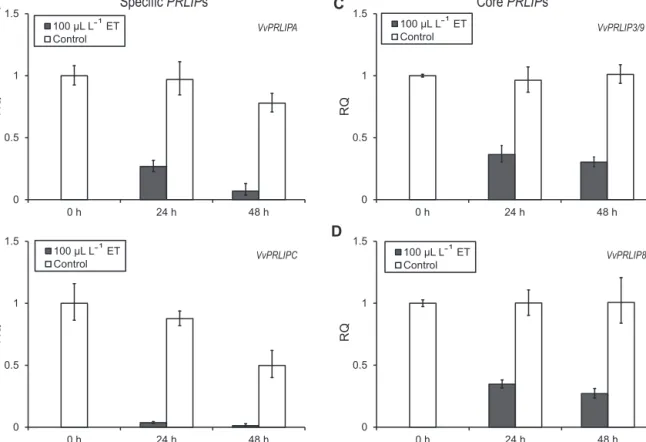
WealsoanalyzedvinestocksofthecultivarPinotNoirtreated with the stress inducers BTH and ET. Increasing expression of bothVvPRLIPAandVvPRLIPCgeneswereobservedinresponseto theSAanalogBTHtreatment(Fig.5AandB),whiletranscription of corePRLIPswasalmost unaffected(Fig.5C and D). ETtreat- mentrepressedtranscriptionofalltheVvPRLIPs(Fig.6),especially VvPRLIPA,toayieldof0.1-foldafter48handVvPRLIPC,whichwas practicallyundetectableatthetranscriptlevelafter2daysofinoc- ulation(Fig.6AandB).Theefficiencyofthechemicaltreatments wasverifiedbymarkergenesVvHSR1andVvPR1accordingtoChong etal.(2008);seeSupplementaryFig.3.
Discussion
ThePRLIPfamilyisayetunclassifiedgroupofinducibledefense relatedgenesresemblinginductioncharacteristicsofgenesencod- ing typical PRproteins. It consistsof 9 paralogs in A. thaliana.
Theprimarydescriptionofthegenesstressedtheinducibilityof PRLIP1andPRLIP2inresponsetoSAtreatmentandPseudomonas infectionandanupregulationofPRLIP6toMeJAtreatment(Jakab etal.,2003).In contrast,otherfamilymembers(namelyPRLIP3, PRLIP8andPRLIP9genes)showconstitutiveexpressionwithonly
0 100 200 300 400 500 600
0 5 10 15 20 25
Control 5-10% 30-40% >75%
RQ
VvPRLIPA VvPRLIPC
0 0.5 1 1.5 2
0 0.5 1 1.5 2
Control 5-10% 30-40% >75%
RQ
VvPRLIP3/9 VvPRLIP8
0 20 40 60 80 100 120 140
0 20 40 60 80
Control 5-10% 30-40% >75%
RQ
B
VvPRLIPA VvPRLIPC
0 0.5 1 1.5 2 2.5
0 0.5 1 1.5 2 2.5
Control 5-10% 30-40% >75%
RQ
D
VvPRLIP3/9
VvPRLIP8
Fig.4.RelativeexpressionpatternsofgrapevinePRLIPgenesuponpowderymildewinfection.LeftscalesindicateexpressionchangesofVvPRLIPA(AandB)andVvPRLIP3/9(C andD),rightscalesindicateVvPRLIPC(AandB)andVvPRLIP8(CandD).AandCrepresentcultivarPinotNoir,whileBandDshowthecultivarCabernetSauvignon.Percentages indicatetherelativeareaoftheinfectedleafsurface,0:samplefromuninfectedleaves.Errorbarscorrespondtotechnicalreplicates.
Specific PRLIP s Core PRLIP s
0 1 2 3 4 5 6 7
0 h 48 h 96 h
RQ
VvPRLIPA 600 mM BTH
Control
A
0 1 2 3 4 5 6 7
0 h 48 h 96 h
RQ
VvPRLIP3/9 600 mM BTH
Control
C
0 1 2 3 4 5 6 7
0 h 48 h 96 h
RQ
VvPRLIPC 600 mM BTH
Control
B
0 1 2 3 4 5 6 7
0 h 48 h 96 h
RQ
VvPRLIP8 600 mM BTH
Control
D
Fig.5.EffectofBTHtreatmentongrapevinePRLIPtranscriptionlevelsinthePinotNoircultivar.Timepointsindicatethehourstheplantsweresampledafterthetreatment.
Errorbarscorrespondtotechnicalreplicates.
Specific PRLIP s Core PRLIP s
0 0.5 1 1.5
0 h 24 h 48 h
RQ
VvPRLIPA 100 μL Lˉ ET¹
Control
100 μL Lˉ ET¹ Control
100 μL Lˉ ET¹ Control 100 μL Lˉ ET¹
Control
A
0 0.5 1 1.5
0 h 24 h 48 h
RQ
VvPRLIP3/9
C
0 0.5 1 1.5
0 h 24 h 48 h
RQ
VvPRLIPC
B
0 0.5 1 1.5
0 h 24 h 48 h
RQ
VvPRLIP8
D
Fig.6. TranscriptionalchangesofVvPRLIPsfollowedbyETtreatmentinthecultivarPinotNoir.Timepointsindicatethehourstheplantsweresampledafterthetreatment.
Errorbarscorrespondtotechnicalreplicates.
minordifferences under stress conditions.In thepresent stud- ies, we screened sequenced plant genome databases to collect PRLIP protein homologs. A phylogenetic tree constructed from theseproteinsindicatesthatangiospermPRLIPhomologsformtwo sisterclades. CladeIis a largegroupof sequences, alldisplay- inghomologytotheArabidopsisPRLIP8sequence,whileCladeII iscomprisedbyhomologs oftheArabidopsisPRLIP3andPRLIP9 andseveraladditionalsubcladesoftaxon-specificPRLIPparalogs.
Eachofthesesubcladesoriginatesfromadefinedplantgenome andthegenesencodingthesetaxon-specificproteinsaregener- allytightlyclustered.TheAtPRLIP1,AtPRLIP2andAtPRLIP6genes showingdefense-relatedcharacteristicsallbelongtooneofthese specificparaloggroups,and arethusuniquefor theArabidopsis genome.Altogether,we termed theseproteinsgenome-specific PRLIPs.TotestourhypothesisthatthesespecificPRLIPsevolved toserveplant–pathogenresponses,wealsoanalyzedexpression patternsofthegenome-specificPRLIPparalogsofgrapevine.The expressionratioofthetwoVvPRLIPgenesVvPRLIPAandVvPRLIPC increasedstronglyuponpowderymildewinfectioninbothPinot NoirandCabernetSauvignoncultivars.TreatmentwithBTHalso increasedtheexpressionoftheseVitis-specificPRLIPgenes,butto amuchlesserextentthanuponpowderymildewinfection.Inter- estingly,although specificPRLIPgenesof thesespeciespossibly evolvedindependently(likelyasaresultoftandemgenedupli- cation),bothAtPRLIP1andAtPRLIP2genesofArabidopsis,justas VvPRLIPAandVvPRLIPCgenesofgrapevine,areallupregulatedby BTHand bytheSAR-inducingpathogens P.syringae orErysiphe necator,respectively.On theotherhand,dissimilaritiescanalso beestablishedamongspecificparalogs.Elevatedtranscriptionof AtPRLIP1couldbeobservedinET-treatedplants,whilethesame treatmentgreatlydecreasedmRNAlevelsofallVvPRLIPs,especially VvPRLIPCandVvPRLIPAtranscription.
Recenttandemduplicationeventscontributingtoplantdefense seem to be a plausible explanation for the high level of
homologyamongclusteredPRLIPgeneswithinacertaingenome, butthepossibilityofconcertedevolutionmight bealsoconsid- ered(Liao,1999),aswasdemonstratedforthePR-10family(Lebel etal.,2010).However,thepresenceofnon-functionalparalogsin bothVitisandArabidopsisreferstoindependentevolutionofdupli- catedgenesundergoingpurifyingselectionasproposedbyNeiand Rooney(2005).
AtPRLIP8,AtPRLIP3 and itsadditional paralog(inArabidopsis) AtPRLIP9 werealsostudiedin detailand werefoundtodisplay significantbasal expression in allstudied organs of Arabidopsis (SzalontaiandJakab,2010).AsbothPRLIP8andPRLIP3/9seemto occurcommonlyinallplants,wetermedthemcorePRLIPsinthe presentstudy.BothAtPRLIP3andAtPRLIP9werealsoinducibleby NaCl,andtheformerbyJAaswell,buttheseexpressionchanges weregenerallylowerthanstressresponsesofthegenome-specific ArabidopsisPRLIPs.AtPRLIP3displayedonlyabrieftransientinduc- tioninresponsetoSAtreatmentinArabidopsis.Thiswasalsotrue forthegrapevineorthologVvPRLIP3/9expression,whichwasonly moderatelyaffectedbyanyofthestressconditionsapplied.How- ever,neitherAtPRLIP8norVvPRLIP8islikelytobeconnectedtoany stressresponse,asnoneofthesegenesdisplayedmarkedchanges in mRNA levels in response to pathogen infection or chemical treatments.Rangesoforgan-specificexpressiondifferenceswere alsogenerallylowincorePRLIPhomologs,especiallyinthecase of AtPRLIP8in Arabidopsisand both VvPRLIP3/9and VvPRLIP8in grapevine(Jakabetal.,2003;SzalontaiandJakab,2010).
Takentogether,wesuggestthat,inadditiontothecorePRLIPs thatarepresentinallanalyzedplanttaxa,genome-specificgroups, whichcontainuniquePRLIPparalogproteinsusuallyencodedin agenecluster,canbeobservedinseveralplanttaxa.Thesegene clustersincludePRLIP1,PRLIP2,PRLIP4,PRLIP5,PRLIP6andPRLIP7in ArabidopsisandVvPRLIPE,VvPRLIPA,VvPRLIPF,VvPRLIPC,VvPRLIPG, VvPRLIPD and VvPRLIPB inVitis vinifera. In contrasttogenome- specificPRLIPs,thecorePRLIPs(PRLIP8andPRLIP3/9orthologs)are
pretedasPR-likeparalogs(PRLs)accordingtoVanLoon’soriginal nomenclature(VanLoonetal.,2006)andmightservesomeother biochemicalfunctioninhealthyplants.Asimilarphenomenonwas describedinthecaseofthreonindeaminasesoftomato,wherethe geneoftheoriginalTD1enzymeisinvolvedinprimarymetabolism, whileaduplicatedparalogTD2(whichonlyappeared inclosely relatedSolanumspecies)hasevolvedtoprotectplants fromthe herbivoreLepidopteras(Gonzales-Vigiletal.,2011).
Withrespecttothegenome-specificPRLIPgenes,ourfindings suggestthatseveralcharacteristicsshare similaritieswithother genescodingfordifferenttypesofPRproteins.First,membersof thesamePRfamily displayextremediversity intranscriptional inducibility.Forinstance,thereisonlyoneArabidopsisPR-1para- loggeneshowingupregulationviabiotrophicpathogenattackorSA treatment.Severalothermembers,however,cannotberelatedto stressresponses(VanLoonetal.,2006).Conversely,ricePR-1genes areinducedbydifferentstressstimuli(Mitsuharaetal.,2008),each havingacharacteristicexpressionpattern.NoneoftheapplePR- 1genescouldyet berelated toanystress responses(Bonasera etal.,2006).Asdiscussedabove,thePRLIPfamilyofArabidopsis andgrapevinecanalsobedividedintomembersclosely related toplant–pathogeninteractions(thegenomespecificPRLIPs)and thosewhicharenot(thecorePRLIPs).Interestingly,theArabidop- sisPRLIP1geneisupregulatedbyantagonisticsignalingpathways mediatedbyETandSA(Jakabetal.,2003)asitisthecaseforsome membersofricePR-1family,especiallyOsPR1a(Mitsuharaetal., 2008).Bycontrast,Vitis-specificPRLIPsareinduciblebyBTHbutare repressedbyET.Moreover,Arabidopsis-specificAtPRLIP6(locatedin thesameclusterasPRLIP1andPRLIP2)isinduciblebyMeJAandET butnotbySAorBTH(Jakabetal.,2003).
Anotheraspectofgeneexpressionvariationistheorgan-specific anddevelopmentallyregulatedtranscriptioninuntreatedplants.
ThishasalsobeenobservedintypicalPRfamilieslikethethaumatin superfamily,inadditiontoinducibilitybyseveralenvironmental stresses(Liuetal.,2010).Differentialaccumulationoflipidtransfer protein(LTP)transcriptsinresponsetopathogenattack,aswellas organ-specifictranscriptionpatterns,havealsobeendescribedin pepper(Jungetal.,2003).Thioninvariantsofthecerealswheatand ricewerealsoreportedtoberegulatedinatissuespecificmanner (Stec,2006),justlikePRLIPgenesinhealthyorgansofArabidop- sisandgrapevineincludingroot–(PRLIP6,PRLIP3,PRLIP4),silique –(PRLIP2,PRLIP9)orleaf–(PRLIP1,VvPRLIPA,VvPRLIPC)-specific members.
Anothernotable analogybetweenPRLIPsandsomeotherPR families is the gene organization. We show that the genome- specificPRLIPgroupsofdifferentplantsmostlyoccurinonesingle geneclustereach.Similarly,orthologgroupsoftandemlyrepeated genes (displaying differentialexpression) encoding germin-like proteinswere also described from barley(Zimmermann et al., 2006), aswellasPR-1genesofgrapevine,fromwhich14genes outof21formonecluster(Lietal.,2011).TandemarraysofPR-1 genesarepresentin theArabidopsisand Oryzagenome,includ- inggenesmorehomologoustoeachotherthantootherparalogs (VanLoonetal.,2006).Theseclustersoftencontainpseudogenes aswell.Thissuggestssmall-scaleduplicationeventsduringevolu- tionasdescribedbyLebeletal.(2010)inthecharacterizationof thegrapevinePR-10family.
ComparisonofclusterscontainingthespecificPRLIPgenesof twocloselyrelatedArabidopsisspeciesindicatedsmallscaleevolu- tionaryeventsresultingindifferencesincludingtheinequalityin copynumberofPRLIP4likegenes.Thisis,however,opentodebate, sinceacomparativestudybetweenA.thalianaandA.lyratarevealed thatmanyofthetransposedgeneswereflankedbyrepeatedgene sequences (Woodhouse et al., 2010). A previous report onthe
speciesand amongproteinssharingsimilarbiological functions (Tiffin,2004)andtherefore,capableofgeneratingsuchinterspe- cificdifferences.Anotherreportshowedthattandemduplicated genesshowincreasedvariationingeneexpressionwhencompared tosegmentallyduplicatedoruniquegenes(Kliebenstein,2008).
Divergenceinregulationoftranscriptionaftergeneamplification could occur especially in the Arabidopsis-specific PRLIP cluster, whereeachofthegeneshasacharacteristicbasalexpressionpat- ternanddifferentialinducibilitybyvariouskindsofstressstimuli (Jakabetal.,2003).Thismightwellprovideamechanismoffine tuningstressresponses.
ThethirdinterestingcommonfeatureamongPRLIPsandsome otherPRgenesistheappearanceofgenome-specificgroups,which occurred in different plant lineages. Rice- or monocot-specific groupsofclassIIIperoxidaseswereshowntobeencodedbylarge numbersofparaloggroupsmostlylocatedingeneclusters(Passardi etal.,2004),similartotaxon-specificendoglucanasesclusteredin theArabidopsisgenome(Libertinietal.,2004).Geneclustersencod- ingLTPswerealsoproposedasbeingindependentlyexpandedin differentplantphyla(Vignolsetal.,1997).WhencomparingPR- 1genesofArabidopsisandrice,itisstrikingthatonlyoneofthe numerousparalogsislikelytobepresentinbothspecies(andcon- sideredtobeanorthologpairofgenes),andtheothergeneshave evolvedafterthedivergenceofthemonocot/eudicotlineages(Van Loonetal.,2006).SimilarlytothespecificPRLIPs,genesofdefensin likeproteinsalsooccuringeneclusters–theresultsoflocaldupli- cationevents–andcanbeclassifiedintotaxon-ortissue-specific subgroupsinhigherplants(Silversteinetal.,2005).Ouranalyses suggestthatsimilardiversificationofPRLIPsresultedinspecific paraloggroupsconferringdefense.Itisalsoimportanttonotethat notallofthestudiedplantgenomespossesanindividualgenome- specificPRLIP group,sothis mechanism islikely tooccuronly occasionally.However,this isnotunusual,asnotallfamiliesof inducibledefense-relatedproteinsseemtobepresentinallplant species(VanLoonetal.,2006).Forinstance,Bowman–Birktype proteinaseinhibitorscanonlybefoundinthePoaceaeandFabaceae families(Melloetal.,2003).Inconclusion,thepresentanalysisof thegrapevinePRLIPgenefamilyentirelysupportstheformerresults obtainedfromArabidopsis.The emergingpictureshows a novel familyencodinginducibledefense-relatedproteinswithregardto pathogeninducibilityanddifferentialbasalgeneexpression.Taken together,ourdataprovideimportantcluesforfurtherexperiments, andespeciallyforproteinlevelcharacterization–essentialtoverify PRcandidacyandphysiologicalrolesofthegenefamily.
Acknowledgements
WethankDr.KrisztinaSzabadfiforhelpfulcomments.Thiswork wassupportedbySROP4.2.2/B-10/1-2010-0029oftheEU,theHun- garianScientificResearchFoundation(OTKAK101430)andbythe SCOPESprogram(projectIZ73Z0128031).
AppendixA. Supplementarydata
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/
j.jplph.2012.07.010.
References
BonaseraJM,KimJF,BeerSV.PRgenesofapple:identificationandexpressionin responsetoelicitorsandinoculationwithErwiniaamylovora.BMCPlantBiol 2006;6:23.
ChongJ,LeHenanffG,BertschC,WalterB.Identificationexpressionanalysisand characterizationofdefenseandsignalinggenesinVitisvinifera.PlantPhysiol Biochem2008;46(4):469–81.
FalkA,FeysBJ,FrostLN,JonesJDG,DanielsMJ,ParkerJE.EDS1,anessentialcom- ponentofRgene-mediateddiseaseresistanceinArabidopsishashomologyto eukaryoticlipases.ProcNatlAcadSciUSA1999;96(6):3292–7.
FarmerEE,RyanCA.Octadecanoidprecursorsofjasmonicacidactivatethesynthesis ofwound-inducibleproteinaseinhibitors.PlantCell1992;4(2):129–34.
Gonzales-VigilE,BianchettiCM,PhillipsJrGN,HoweGA.Adaptiveevolutionofthre- oninedeaminaseinplantdefenseagainstinsectherbivores.ProcNatlAcadSci USA2011;108(14):5897–902.
Hamiduzzaman MM, Jakab G, Barnavon L, Neuhaus JM, Mauch-Mani B.
-Aminobutyricacid-inducedresistanceagainstdownymildewingrapevine actsthroughthepotentiationofcalloseformationandjasmonicacidsignaling.
MolPlantMicrobeInteract2005;18(8):819–29.
HongJK,ChoiHW,HwangIS,KimDS,KimNH,ChoiduS,etal.Functionofanovel GDSL-typepepperlipasegene,CaGLIP1,indiseasesusceptibilityandabiotic stresstolerance.Planta2008;227(3):539–58.
JakabG,ManriqueA,ZimmerliL,MétrauxJP,Mauch-ManiB.Molecularcharacteri- zationofanovellipase-likepathogen-induciblegenefamilyofArabidopsis.Plant Physiol2003;132(4):2230–9.
JungHW,KimW,HwangBK.Threepathogen-induciblegenesencodinglipidtrans- ferproteinfrompepperaredifferentiallyactivatedbypathogens,abiotic,and environmentalstresses.PlantCellEnviron2003;26(6):915–28.
KimKJ,LimJH,KimMJ,KimT,ChungHM,PaekK.H.GDA-lipase1(CaGL1)contributes towoundstressresistancebymodulationofCaPR-4expressioninhotpepper.
BiochemBiophysResCommun2008;374(4):693–8.
KliebensteinDJ.Aroleforgeneduplicationandnaturalvariationofgeneexpression intheevolutionofmetabolism.PLoSOne2008;3(3):e1838.
KumarD,KlessigDF.High-affinitysalicylicacid-bindingprotein2isrequiredfor plantinnateimmunityandhassalicylicacid-stimulatedlipaseactivity.Proc NatlAcadSciUSA2003;100(26):16101–6.
KwonSJ,JinHC,LeeS,NamMH,ChungJH,KwonSI,etal.GDSLlipase-like1regulates systemicresistanceassociatedwithethylenesignalinginArabidopsis.PlantJ 2009;58(2):235–45.
LebelS,SchellenbaumP,WalterB,MaillotP.CharacterisationoftheVitisvinifera PR10multigenefamily.BMCPlantBiol2010;10:184.
LeeDS,KimBK,KwonSJ,JinHC,ParkOK.ArabidopsisGDSLlipase2playsarolein pathogendefensevianegativeregulationofauxinsignaling.BiochemBiophys ResCommun2009;379(4):1038–42.
LeeKA,ChoTJ.Characterizationofasalicylicacid-andpathogen-inducedlipase-like geneinChinesecabbage.JBiochemMolBiol2003;36(5):433–41.
LiZT,DhekneySA,GrayDJ.PR-1genefamilyofgrapevine:auniquelyduplicatedPR-1 genefromaVitisinterspecifichybridconfershighlevelresistancetobacterial diseaseintransgenictobacco.PlantCellRep2011;30(1):1–11.
LiaoD.Concertedevolution:molecularmechanismandbiologicalimplications.Am JHumGenet1999;64(1):24–30.
Libertini E, Li Y, McQueen-Mason SJ. Phylogenetic analysis of the plant endo-beta-1,4-glucanasegenefamily.JMolEvol2004;5:506–15.
LiuJJ,SturrockR,EkramoddoullahAK.Thesuperfamilyofthaumatin-likeproteins:
itsorigin,evolution,andexpressiontowardsbiologicalfunction.PlantCellRep 2010;29(5):419–36.
LysakMA,BerrA,PecinkaA,SchmidtR,McBreenK,SchubertI.Mechanismsof chromosomenumberreductioninArabidopsisthalianaandrelatedBrassicaceae species.ProcNatlAcadSciUSA2006;103(13):5224–9.
MelloMO,TanakaAS,Silva-FilhoMC.MolecularevolutionofBowman–Birktype proteinaseinhibitorsinfloweringplants.MolPhylogenetEvol2003;27(1):
103–12.
MitsuharaI,IwaiT,SeoS,YanagawaY,KawahigasiH,HiroseS,etal.Characteristic expressionoftwelvericePR1familygenesinresponsetopathogeninfec- tion,wounding,anddefense-relatedsignalcompounds(121/180).MolGenet Genomics2008;279(4):415–27.
NeiM,RooneyAP.Concertedandbirth-and-deathevolutionofmultigenefamilies.
AnnuRevGenet2005;39:121–52.
OhIS,ParkAR,BaeMS,KwonSJ,KimYS,LeeJE,etal.Secretomeanalysisrevealsan ArabidopsislipaseinvolvedindefenseagainstAlternariabrassicicola.PlantCell 2005;17(10):2832–47.
OllisDL,CheahE,CyglerM,DijkstraB,FrolowF,FrankenSM,etal.Thealpha/beta hydrolasefold.ProteinEng1992;5(3):197–211.
PassardiF,LongetD,PenelC,DunandC.TheclassIIIperoxidasemultigenicfamily inriceanditsevolutioninlandplants.Phytochemistry2004;65(13):1879–93.
PfafflMW.Anewmathematicalmodelforrelativequantificationinreal-time RT-PCR.NucleicAcidsRes2001;29(9):e45.
Riemann M,GutjahrC, Korte A, RiemannM,Danger B, Muramatsu T,et al.
GER1, aGDSL motif-encoding genefrom rice is a novel earlylight- and jasmonate-inducedgene.PlantBiol(Stuttg)2007;9(1):32–40.
SambrookJ,FritschEF,ManiatisT.Molecular,cloningalaboratorymanual.2nded.
ColdSpringHarbor,NY,USA:ColdSpringHarborLaboratoryPress;1989.
ShahJ.Lipids,lipases,andlipid-modifyingenzymesinplantdiseaseresistance.Annu RevPhytopathol2005;43:229–60.
Silverstein KA, Graham MA, Paape TD, VandenBosch KA. Genome organiza- tion of more than 300 defensin-like genes in Arabidopsis. Plant Physiol 2005;138(2):600–10.
Stec B. Plant thionins – the structural perspective. Cell Mol Life Sci 2006;63(12):1370–85.
SwoffordDL.PAUP*.Phylogeneticanalysisusingparsimony(*andothermethods).
Version4.Sunderland,Massachusetts:SinauerAssociates;2003.
SzalontaiB,JakabG.DifferentialexpressionofPRLIPs,apathogenesis-relatedgene familyencodingclass3lipase-likeproteinsinArabidopsis. ActaBiolHung 2010;61(Suppl.):156–71.
TiffinP.ComparativeevolutionaryhistoriesofchitinasegenesinthegenusZeaand familyPoaceae.Genetics2004;167(3):1331–40.
VanLoonLC,RepM,PieterseCM.Significanceofinducibledefense-relatedproteins ininfectedplants.AnnuRevPhytopathol2006;44:135–62.
VignolsF,WiggerM,García-GarridoJM,GrelletF,KaderJC,DelsenyM.Ricelipid transferprotein(LTP)genesbelongtoacomplexmultigenefamilyandare differentiallyregulated.Gene1997;195(2):177–86.
VlotAC,LiuPP,CameronRK,ParkSW,YangY,KumarD,etal.Identificationoflikely orthologsoftobaccosalicylicacid-bindingprotein2andtheirroleinsystemic acquiredresistanceinArabidopsisthaliana.PlantJ2008;56(3):445–56.
WiermerM,FeysBJ,ParkerJE.Plantimmunity:theEDS1regulatorynode.CurrOpin PlantBiol2005;8(4):383–9.
WoodhouseMR,PedersenB,FreelingM.TransposedgenesinArabidopsisareoften associatedwithflankingrepeats.PLoSGenet2010;6(5):e1000949.
ZhouN,TootleTL,TsuiF,KlessigDF,GlazebrookJ. PAD4functionsupstream from salicylic acidto control defense responses inArabidopsis. Plant Cell 1998;10(6):1021–30.
ZimmermannG,BäumleinH,MockHP,HimmelbachA,SchweizerP.Themultigene familyencodinggermin-likeproteinsofbarley.RegulationandfunctioninBasal hostresistance.PlantPhysiol2006;142(1):181–92.