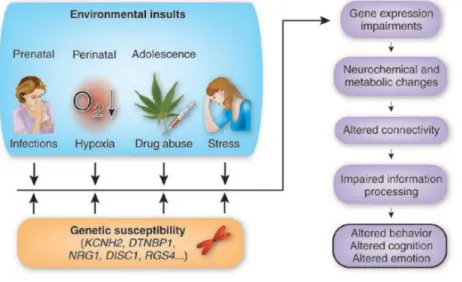
Some of these changes were also present in resting subjects without medication (Keshavan et al., 1998). However, a number of studies failed to replicate the reported differences (Highley et al., 2001; Kulynych et al., 1996).
AIMS
Understanding changes in the gene expression patterns in the postmortem brains of subject with schizophrenia using an unbiased, hypothesis-free, data-driven
Verifying the newly discovered gene expression changes
Investigating the relationships between gene expression changes and genetic susceptibility to schizophrenia
Deciphering the role of the gene expression changes in relationship to pathophysiology and clinical presentation of schizophrenia
Postmortem samples and subjects
Prefrontal cortex was identified based on histological criteria as previously described (Glantz and Lewis, 2000; Volk et al., 2000). Consensus DSM-IV diagnoses were made for all subjects using data from clinical records, toxicology studies, and structured interviews with surviving family members as previously described (Pierri et al., 1999).
DNA Microarrays
The ~1,800 gene expression probes were selected based on 1) previous microarray studies performed in our laboratory, 2) previously published gene expression and genetic data in schizophrenia, and 3) genes participating in cellular processes of implicated in the pathophysiology of PFC dysfunction in schizophrenia. Moreover, based on previous microarray studies of subjects with schizophrenia and matched controls, an addn.
Microarray data analysis
Each microarray consisted of 5 sub-arrays, resulting in a total of 100 measurements/gene/array (5 probes x 4 replicate spots x 5 identical sub-arrays). As a result, the microarray probe list was enriched with GABA, glutamate, synaptic, glial, dopamine, serotonin system and other transcripts.
In situ hybridization
The methods used for in situ hybridization have been previously published (Campbell et al., 1999), with subject pairs always processed together. The OD of the white matter in the section was subtracted from each of these measurements to give five relative OD values for each subject.
The mean relative OD was calculated for each section in five non-overlapping rectangular regions approximately 500 microns wide and scaled lengthwise to encompass cortical layers 2–6. The efficiency of each primer set was assessed prior to qPCR measurements, and a primer set was valid if its yield was >80%.
Hierarchical clustering
qPCR reactions were performed on an ABI Prism 7300 thermal cycler (Applied Biosystems Inc.), quantified using ABI Prism 7300 SDS software (with automatic baseline and automatic threshold detection options selected), and statistically analyzed using a t- pair of Student test in Microsoft Excel.
Presynaptic gene expression changes in the PFC of subjects with schizophrenia
Schizophrenic subjects differed in the number of genes in the PSYN group that decreased over the 95% confidence level. Assessment of the expression of genes belonging to various metabolic pathways in the PFC of subjects with schizophrenia.
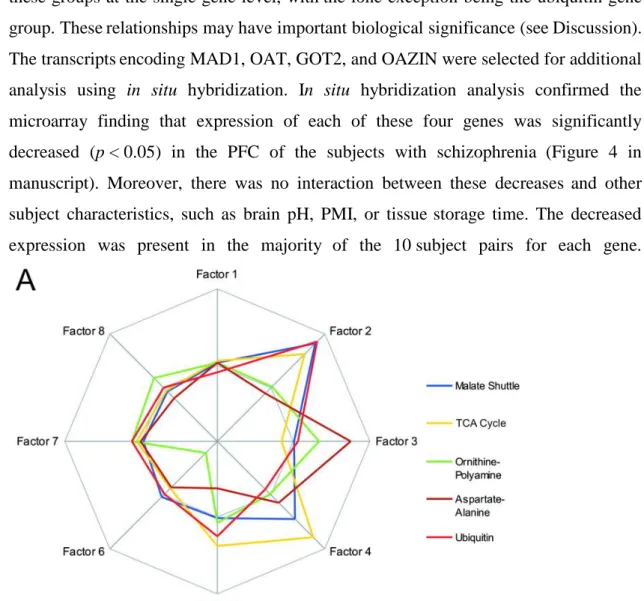
Assessment of the expression of genes belonging to various metabolic pathways in the PFC of subjects with schizophrenia
Most of the metabolic gene clusters we analyzed showed no significant differences in transcript levels between schizophrenic and control subjects (Fig. 1 of the manuscript). However, five gene clusters showed significant changes (p < 0.05) in transcript levels in five or more of the 10 array comparisons.
Altered expression of the 14-3-3 gene family in schizophrenia
Using PCR, we confirmed the robust expression of all seven 14-3-3 genes in area 9 of the human PFC. Although most 14-3-3 genes were highly expressed in the brain, 14-3-3 sigma appeared to be less abundant in this analysis.
Increased expression of immune and chaperone genes in schizophrenia
We examined the expression pattern of the two most strongly affected 14-3-3 genes in human studies (14-3-3 beta and zeta) in three to four sections of each animal. Of the 19 fully annotated transcripts reported to have increased expression in subjects with schizophrenia, 10 (> 50%) were associated with functions in the immune/parallel system (Table 2A and Table 2B in the manuscript).

GABA-ergic expression changes in schizophrenia
The percent expression changes measured by in situ hybridization and qPCR were highly correlated for GAD67. The expression patterns of GAD67, SST, and GABAA α1 mRNAs, as revealed by in situ hybridization, were not different across the groups of monkeys (manuscript figure 6).
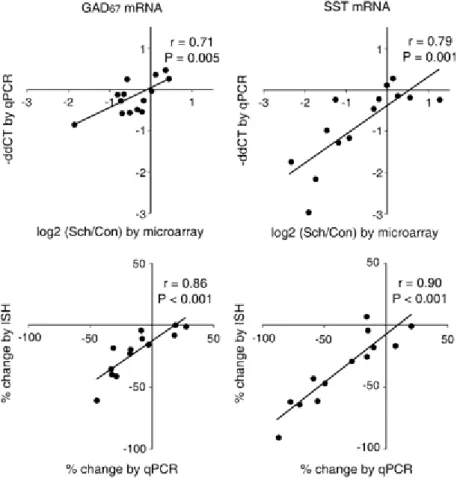
RGS4 – a gene critically involved in schizophrenia?
To verify the microarray findings for the decrease in RGS4 expression, we performed in situ hybridization on the PFC of ten subject pairs of schizophrenic control subjects. Based on optical density analysis, 9/10 subject pairs showed a decrease from 10.2% to 74.3% in PFC RGS4 expression (manuscript Figure 2B). In situ hybridization analysis of PFC region 9 in ten subjects with MDD revealed no difference in mean levels of RGS4 (+3.5%) expression (F1.15=0.27; p=0.61) compared to matched controls (manuscript Figure 2C).
In situ hybridization analysis in four matched pairs of chronically treated and control cynomolgus monkeys showed no difference in PFC RGS4 expression (mean = +5.3%; p = 0.26). Next, to investigate whether there is a more widespread cortical deficiency in the RGS4 transcript, we assessed RGS4 expression in the visual (VC) and motor (MC) cortex of the same 10 pairs of control and schizophrenic subjects by in situ hybridization. . PFC RGS4 expression levels are reduced in nine out of ten subjects with schizophrenia. (a) Emulsion-dipped in situ hybridization micrographs of PFC tissue sections from a schizophrenic (622s) and a matched control (685c), viewed under dark field illumination.
In subjects with schizophrenia, RGS4 expression was consistently reduced in the PFC, VC, and MC (Figure 7 in the manuscript).
DISCUSSION
36. et al., 2000), suggesting that the experimental data support the synaptic-neurological developmental model of schizophrenia that we propose. During childhood, an abundance of excitatory and inhibitory synapses are produced in the cerebral cortex (Bourgeois et al., 1994; Huttenlocher, 1979). Interestingly, the vast majority of measurable metabolic flux in the brain occurs at synapses (Nudo and Masterton, 1986; Sokoloff et al., 1977).
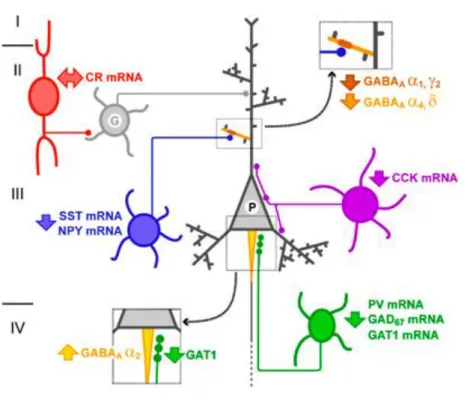
The coupling of GABA(B)R2 to R1 allows surface expression of GABA(B)R1 and the occurrence of potassium and calcium currents (Jones et al., 2000). In mice, mid-pregnancy prenatal exposure to poly(I:C) reduces the number of reelin-positive cells in the hippocampus (Meyer et al., 2006). Because SST- and NPY-containing neurons selectively target distal dendrites of pyramidal neurons (Gonchar et al.), these coordinated gene expression changes suggest that GABA neurotransmission is altered in the dendritic domain of pyramidal neurons in the DLPFC of individuals with schizophrenia.
These changes were associated with the downregulation of GAT1 in the presynaptic terminals of PV-containing chandelier neurons (Pierri et al., 1999) and the upregulation of GABAA.
CONCLUSIONS Major conclusions
Postmortem human brain tissue can be analyzed by high throughput gene expression profiling, and this analysis gives us unique insight into the
It appears that important relationships may exist between altered gene expression and genetic susceptibility to psychiatric disorders (Mirnics and Pevsner,
Lo et al., 2004) show expression changes in the postmortem brain of subjects with schizophrenia, and these genes have also been implicated as putative, inherited schizophrenia susceptibility genes. Thus, we propose that altered expression in the postmortem human brain may have a dual origin for some genes: polymorphisms in the candidate genes themselves or upstream genetic-environmental factors converging to alter their expression level. Based on the currently available data, we hypothesize that certain gene products, functioning as "molecular hubs", commonly show altered expression in psychiatric disorders and confer genetic susceptibility to one or more diseases (Mirnics and Pevsner, 2004; Mirnics et al. .., 2006; Winden et al.. RGS4 appears to be one of the genes critically involved in the pathophysiology of schizophrenia.
It appears that RGS4 is one of the genes critically involved in the pathophysiology of schizophrenia. First, RGS4 mRNA and protein expression are both
The outcome high-throughput gene expression profiling experiments (e.g
Based on the currently available data, we propose the hypothesis that certain gene products acting as “molecular hubs” commonly show altered expression in psychiatric disorders and confer genetic susceptibility to one or more diseases (Mirnics and Pevsner, 2004; Mirnics et al. ., 2006;.
Negative data in the DNA microarray experiments should not be interpreted as an ―absence of gene expression change‖ (Mirnics, 2002; Mirnics, 2006; Mirnics et
Using DNA polymer microarray probes improve sensitivity of transcriptome profiling experiments (Hollingshead et al., 2006)
SUMMARY
BIBLIOGRAPHY
Role of cytokines in mediating effects of prenatal infection on the fetus: implications for schizophrenia. 2004) Microarray analysis of postmortem temporal cortex from patients with schizophrenia. 2004) Interneuron Diversity series: Interneuronal neuropeptides - endogenous regulators of neuronal excitability. Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. 2004) Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia.
2008b) Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Karson C, Griffin W, Mrak R, Sturner W, Shillcutt S, Guggenheim F. 1996) Reduced levels of synaptic proteins in the prefrontal cortex in schizophrenia. Altered 14-3-3 gene expression in the prefrontal cortex of subjects with schizophrenia. validation of microarray results from postmortem brain studies.
Pierri JN, Chaudry AS, Woo TU, Lewis DA. 1999 ) Changes in chandelier neuron axon terminals in the prefrontal cortex of schizophrenic subjects. Reduced somal size of deep layer 3 pyramidal neurons in the prefrontal cortex of subjects with schizophrenia. Volk D, Austin M, Pierri J, Sampson A, Lewis D. 2001) GABA transporter-1 mRNA in the prefrontal cortex in schizophrenia: decreased expression in a subset of neurons.
CANDIDATE’S PUBLICATIONS RELATED TO THESIS
Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA. 2008) Changes in the GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. 2008) Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Mirnics K, Middleton FA, Lewis DA, Levitt P. 2001) Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse.
Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. 2001) Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mirnics K, Korade Z, Arion D, Lazarov O, Unger T, Macioce M, Sabatini M, Terrano D, Douglass KC, Schor NF, Sisodia SS. 2006) Microarrays in brain research: Data quality and limitations revisited. 2006) Critical appraisal of DNA microarrays in psychiatric genomics. Mirnics K, Norstrom EM, Garbett K, Choi SH, Zhang X, Ebert P, Sisodia SS. 2008) Molecular signatures of neurodegeneration in the cortex of PS1/PS2 double knockout mice.
Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Ge Schnell DH. 2009).
CANDIDATE’S PUBLICATIONS UNRELATED TO THESIS
[ PubMed ] Koerber HR, Mirnics K, Kavookjian A. M., Ifa A. R., fi kanneen biroo. (1999) Xiinxala ultrastructural boutons ectopic synaptic fibers afferent jalqabaa naannoo haaromfame irraa ka’an. [ PubMed ] Koerber HR, Mirnics K, Laawsan JJ., fi kanneen biroo. (2006) Pilaastikiin sinaaptikii gaanfa dugda lafee dugda ga’eessotaa keessatti: mul’achuu walitti hidhamiinsa dalagaa haaraa erga narviin naannoo haaromfamee booda. Liintas C, Sacco R, Garbett K, Mirnics K, Militerni R, Bravaccio C, Curatolo P, Manzi B, Schneider C, Melmed R, Eliyaa M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Persico AM.
Mirnics ZK, Caudell E, Gao Y, Kuwahara K, Sakaguchi N, Kurosaki T, Burnside J, Mirnics K, Corey SJ. 2004 ) Microarray analysis of Lyn-deficient B cells reveals germinal center-associated nuclear protein and other genes associated with lymphoid germinal center. Mirnics ZK, Yan C, Portugal C, Kim TW, Saragovi HU, Sisodia SS, Mirnics K, Schor NF. 2005) P75 neurotrophin receptor regulates expression of neural cell adhesion molecule 1. Sabatini MJ, Ebert P, Lewis DA, Levitt P, Cameron JL, Mirnics K. 2007) Amygdala gene expression correlates with social behavior in monkeys experiencing maternal separation. Finally, I must thank all the scientists who have worked for me and with me over the years, too numerous to mention individually.
Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia.